exercise solutions
Last updated: 2018-09-05
workflowr checks: (Click a bullet for more information)-
✔ R Markdown file: up-to-date
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
-
✔ Environment: empty
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
-
✔ Seed:
set.seed(20180820)The command
set.seed(20180820)was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible. -
✔ Session information: recorded
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
-
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.✔ Repository version: 513b4fb
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can usewflow_publishorwflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.Ignored files: Ignored: .DS_Store Ignored: .Rhistory Ignored: .Rproj.user/ Ignored: analysis/.DS_Store Ignored: analysis/assets/ Ignored: data-raw/ Ignored: data/csv/ Ignored: data/raster/ Ignored: data/sf/ Ignored: docs/.DS_Store Untracked files: Untracked: .Rbuildignore Untracked: analysis/mapping.Rmd Unstaged changes: Modified: .gitignore Modified: analysis/_site.yml
Expand here to see past versions:
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | ed98c23 | annakrystalli | 2018-09-04 | Build site. |
| Rmd | 33607af | annakrystalli | 2018-09-04 | workflowr::wflow_publish(“analysis/exercise_solutions.Rmd”) |
| html | 0a20853 | annakrystalli | 2018-09-04 | Build site. |
| Rmd | 6d820c1 | annakrystalli | 2018-09-04 | workflowr::wflow_publish(c(“analysis/exercise_solutions.Rmd”)) |
Vector spatial data
1) add a nose!
Create a nose geometry, combine all the shapes into a single sf and then plot the face.
nose <- st_linestring(x = matrix(c(1, -1, 1, -2), ncol = 2, byrow = T))
sfc <- st_sfc(points, line, nose)
face <- st_sf(data.frame(shape = c("eyes", "mouth", "nose"), geom = sfc))
plot(face)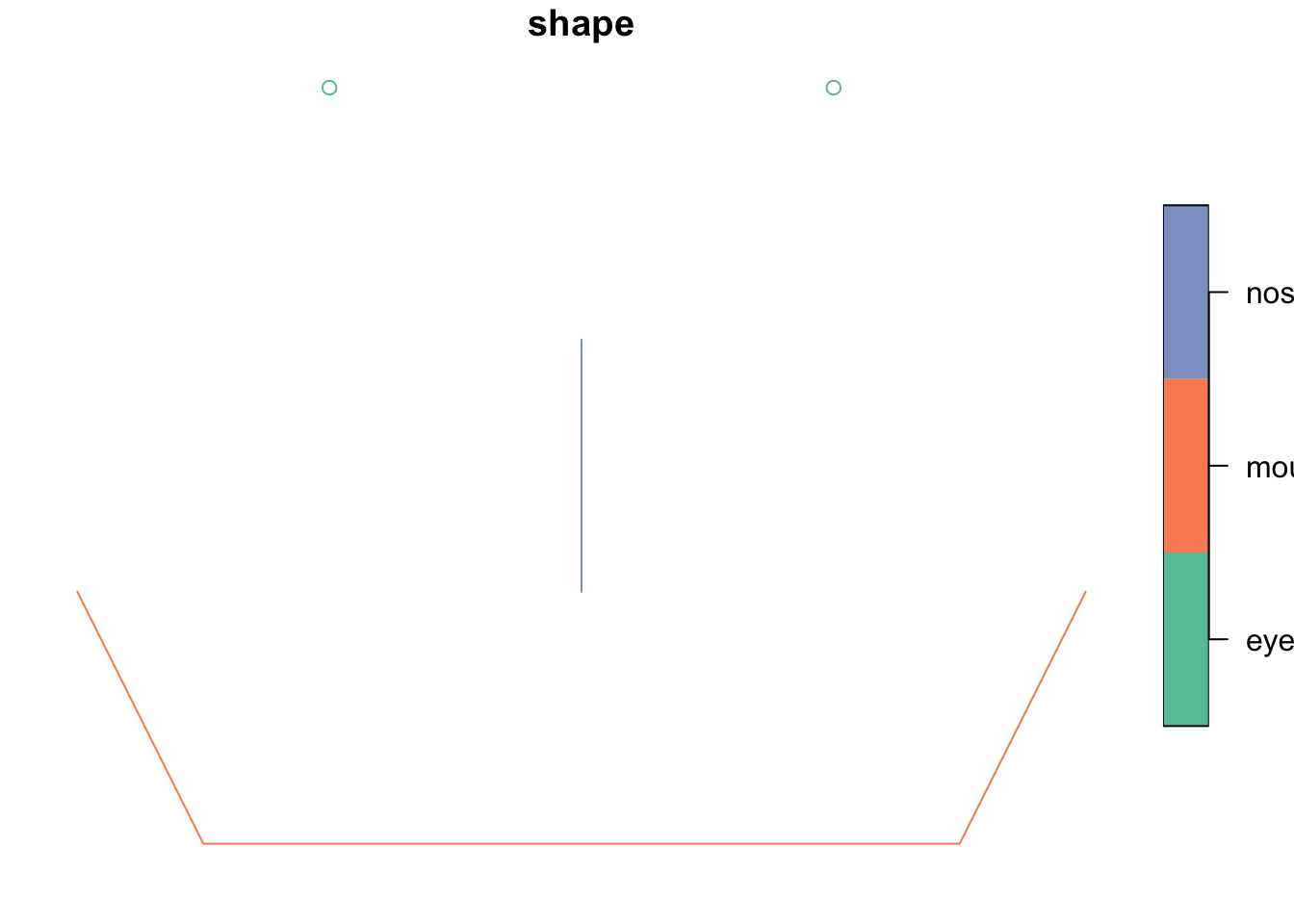
Expand here to see past versions of unnamed-chunk-2-1.png:
| Version | Author | Date |
|---|---|---|
| 0a20853 | annakrystalli | 2018-09-04 |
2) What are the coordinates for the 10th point in the Mexico polygon?
mx_coords <- world %>% filter(iso_a2 == "MX") %>% st_coordinates()
mx_coords[10, c("X", "Y")] X Y
-106.50759 31.75452 3) How about in CRS Mexico ITRF92 / UTM zone 15N
(hint: use st_transform to change the projection first)
mx_utm15 <- world %>%
st_transform(crs = 4488)
mx_coords <- mx_utm15 %>%
filter(iso_a2 == "MX") %>%
st_coordinates()
mx_coords[10, c("X", "Y")] X Y
-784546.6 3593838.9 4) Are these coordinates projected or not? Can you tell by just looking at the CRS in the transformed sf object?
mx_utm15Simple feature collection with 177 features and 10 fields
geometry type: MULTIPOLYGON
dimension: XY
bbox: xmin: -11779910 ymin: -19977280 xmax: 17183260 ymax: 19983850
epsg (SRID): 4488
proj4string: +proj=utm +zone=15 +ellps=GRS80 +towgs84=0,0,0,0,0,0,0 +units=m +no_defs
First 10 features:
iso_a2 name_long continent region_un subregion
1 FJ Fiji Oceania Oceania Melanesia
2 TZ Tanzania Africa Africa Eastern Africa
3 EH Western Sahara Africa Africa Northern Africa
4 CA Canada North America Americas Northern America
5 US United States North America Americas Northern America
6 KZ Kazakhstan Asia Asia Central Asia
7 UZ Uzbekistan Asia Asia Central Asia
8 PG Papua New Guinea Oceania Oceania Melanesia
9 ID Indonesia Asia Asia South-Eastern Asia
10 AR Argentina South America Americas South America
type area_km2 pop lifeExp gdpPercap
1 Sovereign country 19289.97 885806 69.96000 8222.254
2 Sovereign country 932745.79 52234869 64.16300 2402.099
3 Indeterminate 96270.60 NA NA NA
4 Sovereign country 10036042.98 35535348 81.95305 43079.143
5 Country 9510743.74 318622525 78.84146 51921.985
6 Sovereign country 2729810.51 17288285 71.62000 23587.338
7 Sovereign country 461410.26 30757700 71.03900 5370.866
8 Sovereign country 464520.07 7755785 65.23000 3709.082
9 Sovereign country 1819251.33 255131116 68.85600 10003.089
10 Sovereign country 2784468.59 42981515 76.25200 18797.548
geom
1 MULTIPOLYGON (((-11776327 -...
2 MULTIPOLYGON (((7508185 -19...
3 MULTIPOLYGON (((9294103 882...
4 MULTIPOLYGON (((-1664512 58...
5 MULTIPOLYGON (((-1664512 58...
6 MULTIPOLYGON (((473784.4 14...
7 MULTIPOLYGON (((3108708 149...
8 MULTIPOLYGON (((-6664974 -1...
9 MULTIPOLYGON (((-6664974 -1...
10 MULTIPOLYGON (((2134436 -61...We’ve already talked about UTM being projected CRSs but there is also a hint in the proj4string, in particular +units=m indicating that the units are linear (m).
We can also extract the units from an sfs crs using function sf::st_crs() and accessing the units
st_crs(mx_utm15)$units[1] "m"Raster spatial data
1) Create and plot a new rasterLayer of rough mean temperature in degrees C
(rough because it would be much better to use more data at higher temporal resolution, eg at least monthly, not extremes).
We can do this using a simple mean calculation:
rough_mean <- ((full_stack[["mx.bio_5"]] + full_stack[["mx.bio_6"]])/2)/10But we can even use r functions, in this case mean()
rough_mean <- mean(full_stack[["mx.bio_5"]], full_stack[["mx.bio_6"]])/10rough_meanclass : RasterLayer
dimensions : 563, 811, 456593 (nrow, ncol, ncell)
resolution : 0.008333333, 0.008333333 (x, y)
extent : -101.85, -95.09167, 16.94167, 21.63333 (xmin, xmax, ymin, ymax)
coord. ref. : +proj=longlat +datum=WGS84 +no_defs +ellps=WGS84 +towgs84=0,0,0
data source : /Users/Anna/Documents/workflows/workshops/intro-r-gis/data/raster/rough_mean.tif
names : rough_mean
values : -1.45, 29.6 (min, max)levelplot(rough_mean, margin = F)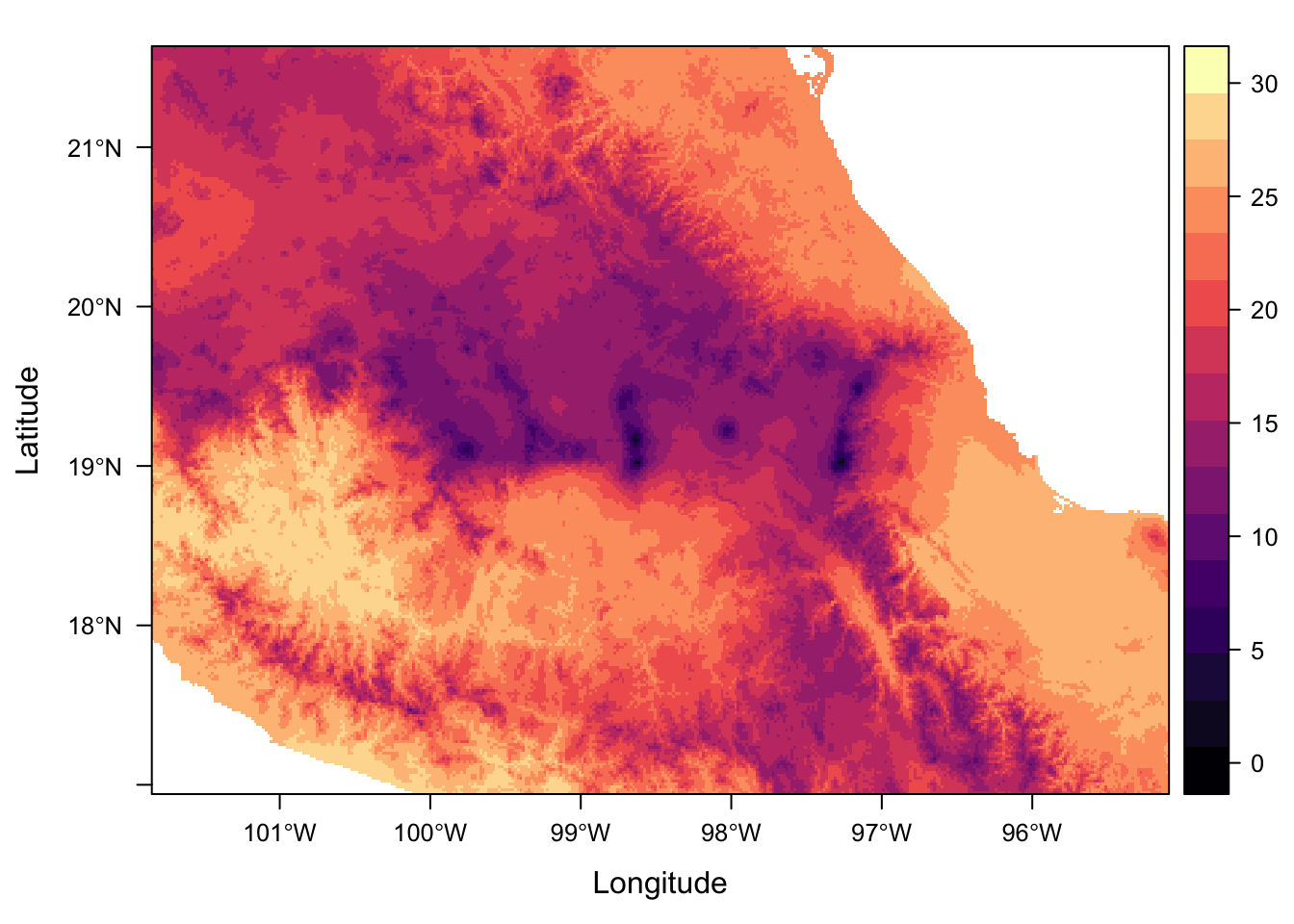
Expand here to see past versions of unnamed-chunk-11-1.png:
| Version | Author | Date |
|---|---|---|
| 0a20853 | annakrystalli | 2018-09-04 |
2) Calculate mean precipitation seasonality for the extraction bounding box area. What is the value?
mean_prec_seasonality <- extract(env_stack[["prec_seasonality"]],
as_Spatial(extract_bbox),
fun = mean,
na.rm = T)
mean_prec_seasonality [,1]
[1,] 87.789573) Add a column indicating whether data points are greater than (TRUE) or less than (FALSE) extraction area mean precipitation seasonality.
mol_env_sf <- mol_env_sf %>%
mutate(greater_mean_ps =
prec_seasonality > as.vector(mean_prec_seasonality))
mol_env_sf %>% select(locality, greater_mean_ps)Simple feature collection with 15 features and 2 fields
geometry type: POINT
dimension: XY
bbox: xmin: -99.84806 ymin: 18.94194 xmax: -97.09056 ymax: 19.63083
epsg (SRID): 4326
proj4string: +proj=longlat +datum=WGS84 +no_defs
# A tibble: 15 x 3
locality greater_mean_ps geometry
<chr> <lgl> <POINT [°]>
1 Nevado de Toluca FALSE (-99.84806 19.19361)
2 Texcalyacac TRUE (-99.5 19.12056)
3 Desierto de los Leones TRUE (-99.30056 19.26667)
4 Ajusco TRUE (-99.3 19.18278)
5 Calpan TRUE (-98.59167 19.13139)
6 Atzompa TRUE (-98.55972 19.18056)
7 Llano Grande FALSE (-98.72056 19.33889)
8 Rio Frio FALSE (-98.69472 19.36611)
9 Nanacamilpa FALSE (-98.59611 19.48028)
10 MalincheS FALSE (-98.02194 19.18722)
11 MalincheW FALSE (-98.095 19.25778)
12 MalincheE FALSE (-97.975 19.23)
13 Texmalaquilla FALSE (-97.29 18.94194)
14 Xometla FALSE (-97.19056 18.975)
15 Vigas FALSE (-97.09056 19.63083)Session information
sessionInfo()R version 3.4.4 (2018-03-15)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS High Sierra 10.13.3
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRlapack.dylib
locale:
[1] en_GB.UTF-8/en_GB.UTF-8/en_GB.UTF-8/C/en_GB.UTF-8/en_GB.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] bindrcpp_0.2.2 spData_0.2.9.3 ggplot2_3.0.0
[4] dplyr_0.7.6 sf_0.6-3 rasterVis_0.45
[7] latticeExtra_0.6-28 RColorBrewer_1.1-2 lattice_0.20-35
[10] raster_2.6-7 sp_1.2-5
loaded via a namespace (and not attached):
[1] zoo_1.8-3 tidyselect_0.2.4 purrr_0.2.5
[4] colorspace_1.3-2 htmltools_0.3.6 viridisLite_0.3.0
[7] yaml_2.1.19 utf8_1.1.3 rlang_0.2.1
[10] R.oo_1.21.0 e1071_1.6-8 hexbin_1.27.1
[13] pillar_1.2.1 glue_1.2.0.9000 withr_2.1.2
[16] DBI_1.0.0 R.utils_2.6.0 bindr_0.1.1
[19] plyr_1.8.4 stringr_1.3.1 munsell_0.5.0
[22] gtable_0.2.0 workflowr_1.0.1 R.methodsS3_1.7.1
[25] evaluate_0.11 knitr_1.20 parallel_3.4.4
[28] class_7.3-14 Rcpp_0.12.18 backports_1.1.2
[31] scales_1.0.0 classInt_0.1-24 digest_0.6.15
[34] stringi_1.2.4 grid_3.4.4 rprojroot_1.3-2
[37] cli_1.0.0 rgdal_1.3-4 here_0.1
[40] tools_3.4.4 magrittr_1.5 lazyeval_0.2.1
[43] tibble_1.4.2 crayon_1.3.4 whisker_0.3-2
[46] pkgconfig_2.0.2 assertthat_0.2.0 rmarkdown_1.10
[49] R6_2.2.2 units_0.6-0 git2r_0.21.0
[52] compiler_3.4.4 This reproducible R Markdown analysis was created with workflowr 1.0.1