Assessing the effects of time series duration
Robert W Schlegel
2019-03-19
Last updated: 2019-03-19
workflowr checks: (Click a bullet for more information)-
✔ R Markdown file: up-to-date
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
-
✔ Environment: empty
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
-
✔ Seed:
set.seed(666)The command
set.seed(666)was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible. -
✔ Session information: recorded
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
-
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.✔ Repository version: 970b22c
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can usewflow_publishorwflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.Ignored files: Ignored: .Rhistory Ignored: .Rproj.user/ Ignored: data/sst_ALL_clim_only.Rdata Ignored: data/sst_ALL_event_aov_tukey.Rdata Ignored: data/sst_ALL_flat.Rdata Ignored: data/sst_ALL_miss.Rdata Ignored: data/sst_ALL_miss_cat_chi.Rdata Ignored: data/sst_ALL_miss_clim_event_cat.Rdata Ignored: data/sst_ALL_miss_clim_only.Rdata Ignored: data/sst_ALL_miss_event_aov_tukey.Rdata Ignored: data/sst_ALL_repl.Rdata Ignored: data/sst_ALL_smooth.Rdata Ignored: data/sst_ALL_smooth_aov_tukey.Rdata Ignored: data/sst_ALL_smooth_event.Rdata Ignored: data/sst_ALL_trend.Rdata Ignored: data/sst_ALL_trend_clim_event_cat.Rdata Untracked files: Untracked: analysis/bibliography.bib Untracked: code/functions.R Untracked: code/workflow.R Untracked: data/.gitignore Untracked: data/clim_py.csv Untracked: data/clim_py_joinAG_1.csv Untracked: data/clim_py_joinAG_5.csv Untracked: data/clim_py_joinAG_no.csv Untracked: data/clim_py_minD_3.csv Untracked: data/clim_py_minD_7.csv Untracked: data/clim_py_pctile_80.csv Untracked: data/clim_py_pctile_95.csv Untracked: data/clim_py_pctile_99.csv Untracked: data/clim_py_random.csv Untracked: data/clim_py_spw_11.csv Untracked: data/clim_py_spw_51.csv Untracked: data/clim_py_spw_no.csv Untracked: data/clim_py_whw_3.csv Untracked: data/clim_py_whw_7.csv Untracked: data/mhwBlock.csv Untracked: data/mhws_py.csv Untracked: data/mhws_py_joinAG_1.csv Untracked: data/mhws_py_joinAG_5.csv Untracked: data/mhws_py_joinAG_no.csv Untracked: data/mhws_py_minD_3.csv Untracked: data/mhws_py_minD_7.csv Untracked: data/mhws_py_pctile_80.csv Untracked: data/mhws_py_pctile_95.csv Untracked: data/mhws_py_pctile_99.csv Untracked: data/mhws_py_random.csv Untracked: data/mhws_py_spw_11.csv Untracked: data/mhws_py_spw_51.csv Untracked: data/mhws_py_spw_no.csv Untracked: data/mhws_py_whw_3.csv Untracked: data/mhws_py_whw_7.csv Untracked: data/sst_ALL.Rdata Untracked: data/sst_ALL_KS.Rdata Untracked: data/sst_ALL_cat_chi.Rdata Untracked: data/sst_ALL_clim_category_count.Rdata Untracked: data/sst_ALL_con.Rdata Untracked: data/sst_ALL_detrend.Rdata Untracked: data/sst_ALL_event_cor.Rdata Untracked: data/sst_ALL_miss_clim_KS_p.Rdata Untracked: data/sst_ALL_miss_clim_aov_p.Rdata Untracked: data/sst_ALL_miss_clim_aov_tukey.Rdata Untracked: data/sst_ALL_miss_event_CI.Rdata Untracked: data/sst_ALL_smooth_R2.Rdata Untracked: data/sst_ALL_smooth_R2_base.Rdata Untracked: data/sst_ALL_smooth_cor_base.Rdata Untracked: data/sst_ALL_smooth_real_category.Rdata Untracked: data/sst_ALL_trend_cat_chi.Rdata Untracked: data/sst_ALL_trend_clim_KS_p.Rdata Untracked: data/sst_ALL_trend_clim_aov_tukey.Rdata Untracked: data/sst_ALL_trend_clim_only.Rdata Untracked: data/sst_ALL_trend_event_CI.Rdata Untracked: data/sst_ALL_trend_event_aov_tukey.Rdata Untracked: data/sst_WA.csv Untracked: data/sst_WA_miss_ice.csv Untracked: data/sst_WA_miss_random.csv Unstaged changes: Deleted: .Rbuildignore Modified: .gitignore Deleted: DESCRIPTION Deleted: NAMESPACE Modified: NEWS.md Deleted: R/MHWdetection-package.R Deleted: R/placeholder.R Modified: README.md Deleted: _pkgdown.yml Modified: _workflowr.yml Modified: analysis/_site.yml Deleted: docs/articles/fig/heatwaveR_v3.svg Deleted: docs/docsearch.css Deleted: docs/docsearch.js Deleted: docs/link.svg Deleted: docs/pkgdown.css Deleted: docs/pkgdown.js Deleted: docs/pkgdown.yml Deleted: docs/sitemap.xml Deleted: man/MHWdetection-package.Rd Deleted: man/placeholder.Rd Deleted: tests/testthat.R Deleted: tests/testthat/test-placeholder.R Deleted: vignettes/.gitignore Deleted: vignettes/Climatologies_and_baselines.Rmd Deleted: vignettes/Short_climatologies.Rmd Deleted: vignettes/best_practices.Rmd Deleted: vignettes/bibliography.bib Deleted: vignettes/data/.gitignore Deleted: vignettes/data/clim_py.csv Deleted: vignettes/data/clim_py_joinAG_1.csv Deleted: vignettes/data/clim_py_joinAG_5.csv Deleted: vignettes/data/clim_py_joinAG_no.csv Deleted: vignettes/data/clim_py_minD_3.csv Deleted: vignettes/data/clim_py_minD_7.csv Deleted: vignettes/data/clim_py_pctile_80.csv Deleted: vignettes/data/clim_py_pctile_95.csv Deleted: vignettes/data/clim_py_pctile_99.csv Deleted: vignettes/data/clim_py_random.csv Deleted: vignettes/data/clim_py_spw_11.csv Deleted: vignettes/data/clim_py_spw_51.csv Deleted: vignettes/data/clim_py_spw_no.csv Deleted: vignettes/data/clim_py_whw_3.csv Deleted: vignettes/data/clim_py_whw_7.csv Deleted: vignettes/data/mhwBlock.csv Deleted: vignettes/data/mhws_py.csv Deleted: vignettes/data/mhws_py_joinAG_1.csv Deleted: vignettes/data/mhws_py_joinAG_5.csv Deleted: vignettes/data/mhws_py_joinAG_no.csv Deleted: vignettes/data/mhws_py_minD_3.csv Deleted: vignettes/data/mhws_py_minD_7.csv Deleted: vignettes/data/mhws_py_pctile_80.csv Deleted: vignettes/data/mhws_py_pctile_95.csv Deleted: vignettes/data/mhws_py_pctile_99.csv Deleted: vignettes/data/mhws_py_random.csv Deleted: vignettes/data/mhws_py_spw_11.csv Deleted: vignettes/data/mhws_py_spw_51.csv Deleted: vignettes/data/mhws_py_spw_no.csv Deleted: vignettes/data/mhws_py_whw_3.csv Deleted: vignettes/data/mhws_py_whw_7.csv Deleted: vignettes/data/sst_ALL.Rdata Deleted: vignettes/data/sst_ALL_KS.Rdata Deleted: vignettes/data/sst_ALL_cat_chi.Rdata Deleted: vignettes/data/sst_ALL_clim_category_count.Rdata Deleted: vignettes/data/sst_ALL_con.Rdata Deleted: vignettes/data/sst_ALL_detrend.Rdata Deleted: vignettes/data/sst_ALL_event_cor.Rdata Deleted: vignettes/data/sst_ALL_miss_clim_KS_p.Rdata Deleted: vignettes/data/sst_ALL_miss_clim_aov_p.Rdata Deleted: vignettes/data/sst_ALL_miss_clim_aov_tukey.Rdata Deleted: vignettes/data/sst_ALL_miss_event_CI.Rdata Deleted: vignettes/data/sst_ALL_miss_event_aov_tukey.Rdata Deleted: vignettes/data/sst_ALL_smooth_R2.Rdata Deleted: vignettes/data/sst_ALL_smooth_R2_base.Rdata Deleted: vignettes/data/sst_ALL_smooth_cor_base.Rdata Deleted: vignettes/data/sst_ALL_smooth_real_category.Rdata Deleted: vignettes/data/sst_ALL_trend_cat_chi.Rdata Deleted: vignettes/data/sst_ALL_trend_clim_KS_p.Rdata Deleted: vignettes/data/sst_ALL_trend_clim_aov_tukey.Rdata Deleted: vignettes/data/sst_ALL_trend_clim_only.Rdata Deleted: vignettes/data/sst_ALL_trend_event_CI.Rdata Deleted: vignettes/data/sst_ALL_trend_event_aov_tukey.Rdata Deleted: vignettes/data/sst_WA.csv Deleted: vignettes/data/sst_WA_miss_ice.csv Deleted: vignettes/data/sst_WA_miss_random.csv Deleted: vignettes/fig/detect_diagram.svg Deleted: vignettes/fig/heatwaveR_v3.svg Deleted: vignettes/gridded_products.Rmd Deleted: vignettes/missing_data.Rmd Deleted: vignettes/r_vs_python.Rmd Deleted: vignettes/r_vs_python_additional.Rmd Deleted: vignettes/r_vs_python_arguments.Rmd Deleted: vignettes/time_series_duration.Rmd Deleted: vignettes/trend.Rmd Deleted: vignettes/variance.Rmd
Expand here to see past versions:
Overview
It has been shown that the length (duration) of a time series may be one of the most important factors in determining the usefulness of those data for any number of applications (Schlegel and Smit 2016). For the detection of MHWs it is recommended that one has at least 30 years of data in order to accurately detect the events therein. This is no longer an issue for most ocean surfaces because many high quality SST products, such as NOAA OISST, are now in exceedence of the 30-year minimum. However, besides issues relating to the coarseness of these older products, there are many other reasons why one would want to detect MHWs in products with shorter records. In situ measurements along the coast are one good example of this, another being the use of the newer higher resolution SST products, a third being the use of reanalysis/model products for the detection of events in 3D. It is therefore necessary to quantify the effects that shorter time series duration (< 30 years) has on the accurate detection of events. Once any discrepancies have been accounted for, best practices must be developed to allow users to improve the precision of the detection of events in their shorter time series.
Time series shortening
A time series derives it’s usefulness from it’s length. This is generally because the greater the number of observations (larger sample size), the more confident one can be about the resultant statistics. This is particularly true for decadal scale measurements and is why the WMO recommends a minimum of 30 years of data before drawing any conclusions about decadal (long-term) trends observed in a time series. We however want to look not at decadal scale trends, but rather at how observed MHWs compare against one another when they have been detected using longer or shorter time series. In order to to quantify this effect we will use the minimum and maximum range of time series available to us. The maximum length will be 37 years, as this is the full extent of the NOAA OISST data included in heatwaveR. The minimum length for comparing the seasonal signal and 90th percentile thresholds will be three years as his is the minimum number of years required to calculate a climatology. For comparing MHWs the minimum will be ten years so that we may still have enough events detected between shortened time series to produce robust statistics.
Before we can discuss shortening techniques, we must first consider the inherent decadal trend in the time series themselves. This is usually the primary driver for much of the event detection changes over time (Oliver et al. 2018). Therefore, if we truly want to understand the effect that a shortened time series may have on event detection, apart from the effect of the decadal trend, we must de-trend the time series first. To this end there is an entire separate vignette that quantifies the effect of long-term trends on the detection of events.
The time series shortening technique proposed here is re-sampling. This methodology takes the full 37 year de-trended time series and randomly samples n years from it to simulate a 3 – 37 year time series. One then uses the jumbled up, randomly selected years of data to create the seasonal signal and 90th percentile threshold (hereafter referred to as “the climatologies”). Re-sampling the time series in this way is useful because it allows us to replicate random climatology creation 100 (or more) times for each time series length in order to produce a more confident estimation of how likely climatologies generated from certain time series lengths are to impact the accuracy of event detection. It also allows us to see how consistent the size of MHWs are in a given time series, or if they very quite a bit depending on the years used.
The WMO recommends that the period used to create a climatology be 30 years in length, starting on the first year of a decade (e.g. 1981), however; because we are going to be running these analyses on many time series shorter than 30 years, for the sake of consistency we will use all of the years present in each re-sampled time series them as the climatology period, regardless of length. For example, if a time series spans 1982 – 2018, that will be the period used for calculating the climatologies. Likewise, should the time series only span the years 2006 – 2018, that will be the period used.
The effect this has on the climatologies will be quantified through the following statistics:
- An ANOVA will compare the climatologies created from different lengths;
- A post-hoc Tukey test to determine where the difference in time series length for climatology creation become significant;
- Correlations between time series length/SD and seas/thresh mean/SD
- Kolmogorov-Smirnov test for where the climatologies diverge significantly from the 30 year climatologies
The effect this has on the MHW metrics will be quantified with the following statistics:
- ANOVA comparing the primary metrics for MHWs detected using different time series lengths;
- A post-hoc Tukey test to show where the difference in time series length for event detection become significant;
- Confidence intervals (CIs) for the measured metrics
- Correlations between time series length/SD and MHW metrics
In addition to checking all of the statistics mentioned above, it is also necessary to record what the overall relationship is with the reduction of time series length and the resultant climatologies/metrics. For example, are MHWs on average more intense in shorter time series or less? We must also look into how the categories of the events are affected. This will form part of the best practice advice later.
Re-sampling
Below we use re-sampling to simulate 100 time series at each length of 3 – 37 years on de-trended data. This is presently being done with the three pre-packaged time series available in the heatwaveR package and the python distribution, but eventually will be run on the global data. The original data are appended to these 100 re-samples as rep = 0.
library(tidyverse)
library(broom)
library(heatwaveR, lib.loc = "~/R-packages/")
# cat(paste0("heatwaveR version = ",packageDescription("heatwaveR")$Version))
library(lubridate) # This is intentionally activated after data.table
library(fasttime)
library(ggpubr)
library(boot)
library(FNN)
library(mgcv)
library(doMC); doMC::registerDoMC(cores = 50)
MHW_colours <- c(
"I Moderate" = "#ffc866",
"II Strong" = "#ff6900",
"III Severe" = "#9e0000",
"IV Extreme" = "#2d0000"
)# First put all of the data together and create a site column
sst_ALL <- rbind(sst_Med, sst_NW_Atl, sst_WA) %>%
mutate(site = rep(c("Med", "NW_Atl", "WA"), each = nrow(sst_WA)))
save(sst_ALL, file = "data/sst_ALL.Rdata")
# Function for creating a re-sample from 37 years of data
sample_37 <- function(rep){
year_filter <- c(1,2,3)
while(length(unique(year_filter)) != 37){
year_filter <- factor(sample(1982:2018, size = 37, replace = F))
}
res <- data.frame()
for(i in 1:length(year_filter)){
df_sub <- sst_ALL %>%
filter(year(t) == year_filter[i],
# Remove leap-days here
paste0(month(t),"-",day(t)) != "2-29") %>%
mutate(year = seq(1982,2018)[i],
month = month(t),
day = day(t),
year_orig = year(t)) %>%
mutate(t = as.Date(fastPOSIXct(paste(year, month, day, sep = "-")))) %>%
select(-year, -month, -day)
res <- rbind(res, df_sub)
}
res$rep <- as.character(rep)
return(res)
}
# Re-sample the data 100 times
doMC::registerDoMC(cores = 50)
sst_ALL_repl <- plyr::ldply(2:100, sample_37, .parallel = T)
# Add the original data as rep = "1"
sst_ALL_1 <- sst_ALL %>%
mutate(year_orig = year(t),
rep = "1")
sst_ALL_repl <- rbind(sst_ALL_1, sst_ALL_repl)
# Save and clear
save(sst_ALL_repl, file = "data/sst_ALL_repl.Rdata")
rm(sst_ALL_1, sst_ALL_repl); gc()# Load randomly sample time series
load("data/sst_ALL_repl.Rdata")
# Function to extract the residuals around the linear trend
# This is the de-trended anomaly time series
detrend <- function(df){
resids <- broom::augment(lm(temp ~ t, df))
res <- df %>%
mutate(temp = temp - resids$.fitted) %>%
select(-year_orig)
return(res)
}
# Create de-trended anomaly time series from all re-samples
doMC::registerDoMC(cores = 50)
sst_ALL_flat <- plyr::ddply(sst_ALL_repl, c("site", "rep"), detrend)
# Save and clear
save(sst_ALL_flat, file = "data/sst_ALL_flat.Rdata")
rm(sst_ALL_repl, sst_ALL_flat); gc()# Load de-trended data
load("data/sst_ALL_flat.Rdata")
# Calculate climatologies, events, and categories on shrinking time series
shrinking_results <- function(year_begin){
res <- sst_ALL_flat %>%
filter(year(t) >= year_begin) %>%
mutate(year_index = 2018-year_begin+1) %>%
group_by(rep, site, year_index) %>%
nest() %>%
mutate(clims = map(data, ts2clm,
climatologyPeriod = c(paste0(year_begin,"-01-01"), "2018-12-31")),
events = map(clims, detect_event),
cats = map(events, category)) %>%
select(-data, -clims)
return(res)
}
# Run this preferably with 35 cores for speed, RAM allowing
doMC::registerDoMC(cores = 35)
sst_ALL_smooth <- plyr::ldply(1982:2016, shrinking_results, .parallel = T)
save(sst_ALL_smooth, file = "data/sst_ALL_smooth.Rdata")
rm(sst_ALL_flat, sst_ALL_smooth); gc()Climatology statistics
We will now see if the 100 re-sampled time series produce significantly different climatologies as they are shortened. If so, we also want to see at what point the climatologies begin to diverge.
load("data/sst_ALL_smooth.Rdata")
# Extract climatology values only
sst_ALL_clim_only <- sst_ALL_smooth %>%
unnest(events) %>%
filter(row_number() %% 2 == 1) %>%
unnest(events) %>%
select(rep:doy, seas:thresh) %>%
unique() %>%
arrange(site, rep, -year_index, doy)
save(sst_ALL_clim_only, file = "data/sst_ALL_clim_only.Rdata")
rm(sst_ALL_smooth, sst_ALL_clim_only); gc()ANOVA p-values
# Run an ANOVA on each metric of the combined event results and get the p-value
# df <- res
aov_p <- function(df){
aov_models <- df[ , -grep("year_index", names(df))] %>%
map(~ aov(.x ~ df$year_index)) %>%
map_dfr(~ broom::tidy(.), .id = 'metric') %>%
mutate(p.value = round(p.value, 4)) %>%
filter(term != "Residuals") %>%
select(metric, p.value)
return(aov_models)
}
# Run an ANOVA on each metric and then a Tukey test
aov_tukey <- function(df){
aov_tukey <- df[ , -grep("year_index", names(df))] %>%
map(~ TukeyHSD(aov(.x ~ df$year_index))) %>%
map_dfr(~ broom::tidy(.), .id = 'metric') %>%
mutate(p.value = round(adj.p.value, 4)) %>%
# filter(term != "Residuals") %>%
select(metric, comparison, adj.p.value) %>%
# filter(adj.p.value <= 0.05) %>%
arrange(metric, adj.p.value)
return(aov_tukey)
}
# Quick wrapper for getting results for ANOVA and Tukey on clims
# df <- sst_ALL_clim_only %>%
# filter(rep == "1", site == "WA") %>%
# select(-rep, -site)
clim_aov_tukey <- function(df){
res <- df %>%
select(year_index, seas, thresh) %>%
mutate(year_index = as.factor(year_index)) %>%
nest() %>%
mutate(aov = map(data, aov_p),
tukey = map(data, aov_tukey)) %>%
select(-data)
return(res)
}# This file is not uploaded to GitHub as it is too large
# One must first run the above code locally to generate and save the file
load("data/sst_ALL_clim_only.Rdata")
doMC::registerDoMC(cores = 50)
sst_ALL_smooth_aov_tukey <- plyr::ddply(sst_ALL_clim_only, c("site", "rep"),
clim_aov_tukey, .parallel = T)
save(sst_ALL_smooth_aov_tukey, file = "data/sst_ALL_smooth_aov_tukey.Rdata")
rm(sst_ALL_smooth, sst_ALL_smooth_aov_tukey); gc()load("data/sst_ALL_smooth_aov_tukey.Rdata")
sst_ALL_smooth_aov <- sst_ALL_smooth_aov_tukey %>%
select(-tukey) %>%
unnest()
label_count_aov_p <- sst_ALL_smooth_aov %>%
group_by(site, metric) %>%
filter(p.value <= 0.05) %>%
summarise(count = n())
knitr::kable(label_count_aov_p,
col.names = c("Time series", "Climatology", "Significantly different"),
caption = "Table 1: This table shows the results of 100 ANOVAs on the real and re-sampled time series with systematically shortened lengths from 37 -- 3 years. Note that the climatologies for the Med time series were never significantly different and so aren't shown here.")| Time series | Climatology | Significantly different |
|---|---|---|
| Med | thresh | 1 |
| NW_Atl | seas | 1 |
| NW_Atl | thresh | 23 |
| WA | seas | 52 |
| WA | thresh | 75 |
rm(sst_ALL_smooth_aov, label_count_aov_p); gc() used (Mb) gc trigger (Mb) max used (Mb)
Ncells 1943162 103.8 3944070 210.7 2441957 130.5
Vcells 4460738 34.1 10146329 77.5 6602844 50.4The above table shows that there are no significant differences in the climatologies for the Mediterannean and relatively few for the North West Atlantic time series. We do however see that of the 100 re-samples for the Western Australia time series, 52 of the seasonal signals and 75 of the thresholds did differ significantly at some point during the shortening.
Post-hoc Tukey test
To get a more precise pictures of where these differences are occurring we will need to look at the post-hoc Tukey test results.
sst_ALL_smooth_tukey <- sst_ALL_smooth_aov_tukey %>%
select(-aov) %>%
unnest()
clim_tukey_sig <- sst_ALL_smooth_tukey %>%
filter(adj.p.value <= 0.05) %>%
separate(comparison, into = c("year_long", "year_short"), sep = "-") %>%
mutate(year_long = as.numeric(year_long),
year_short = as.numeric(year_short)) %>%
mutate(year_dif = year_long - year_short) %>%
# select(-adj.p.value, -rep) %>%
group_by(site, metric, year_long, year_short, year_dif) %>%
summarise(count = n()) %>%
ungroup() %>%
arrange(-year_long, -year_dif) %>%
mutate(year_index = factor(paste0(year_long,"-",year_dif)),
year_index = factor(year_index, levels = unique(year_index)))
# levels(clim_tukey_sig$year_index)
ggplot(clim_tukey_sig, aes(x = year_long, y = year_short)) +
# geom_point() +
# geom_errorbarh(aes(xmin = year_long, xmax = year_short)) +
geom_tile(aes(fill = count)) +
facet_grid(metric~site) +
scale_x_reverse(breaks = seq(3, 36, 3)) +
scale_y_reverse(breaks = seq(4 , 16, 2)) +
scale_fill_viridis_c("count out of \n100 re-samples") +
labs(x = "Length (years) of long time series",
y = "Length (years) of short time series")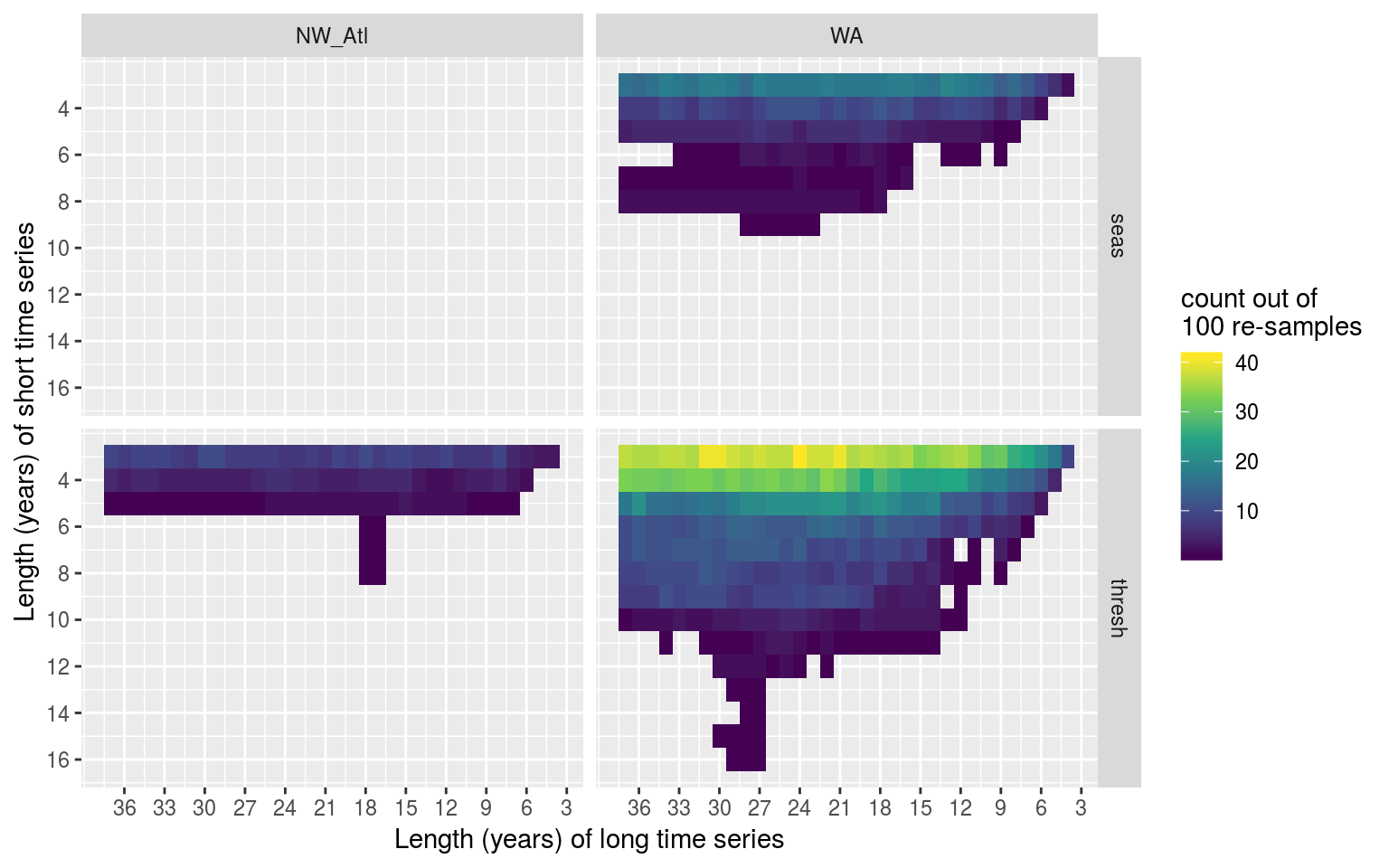
Heatmap showing the distribution of the difference in years for climatologies that were significantly different within the same time series when different lengths of the time series were used to calculate the thresholds. The y axis shows the range of shorter time series lengths that produced significant results when compared to longer time series lengths, which are shown on the x axis. The fill at each pixel shows the count out of 100 re-samples in which the comparison of these two time series lengths were significantly different.
Expand here to see past versions of clim-tukey-heat-1.png:
| Version | Author | Date |
|---|---|---|
| fa7fd57 | robwschlegel | 2019-03-19 |
rm(sst_ALL_smooth_aov_tukey, sst_ALL_smooth_tukey, clim_tukey_sig); gc() used (Mb) gc trigger (Mb) max used (Mb)
Ncells 2080465 111.2 3944070 210.7 3944070 210.7
Vcells 3880214 29.7 10146329 77.5 10146143 77.5The above figure shows us that as few as a four year difference in time series length was able to create significantly different thresholds, but that the most common difference was 14 – 17 years with the distribution left-skewed.
Kolmogorov-Smirnov tests
Whereas the above figure does reveal to us that shorter time series are more often significantly different than longer time series, it takes quie a while to decipher this information and it doesn’t seem very helpful to know this level of detail. Rather I think it would be more useful to simply run a series of Kolmogorov-Smirnov tests for each length of each re-sampled time series to see how often the climatology distributions are significantly different from the climatologies at the 30 year length. This is not only a more appropriate test for these climatology data than an ANOVA, it is also a more simplified way of understanding the results.
load("data/sst_ALL_clim_only.Rdata")
# Function for running a pairwise KS test against the
KS_sub <- function(df_2, df_1){
suppressWarnings( # Suppress warnings about perfect matches
res <- data.frame(seas = round(ks.test(df_1$seas, df_2$seas)$p.value, 4),
thresh = round(ks.test(df_1$thresh, df_2$thresh)$p.value, 4),
year_1 = df_1$year_index[1],
year_2 = df_2$year_index[1])
)
return(res)
}
# Wrapper function to run all pairwise KS tests
# df <- sst_ALL_clim_only %>%
# filter(rep == "30", site == "WA")
KS_all <- function(df){
# The 30 year clims against which all are compared
df_30 <- df %>%
filter(year_index == 30)
# The results
res <- df %>%
mutate(year_index2 = year_index) %>%
group_by(year_index2) %>%
nest() %>%
mutate(KS = map(data, KS_sub, df_1 = df_30)) %>%
select(-data, -year_index2) %>%
unnest() %>%
tidyr::gather(key = "metric", value = "p.value", -year_1, -year_2)
return(res)
}
doMC::registerDoMC(cores = 50)
sst_ALL_KS <- plyr::ddply(sst_ALL_clim_only, KS_all,
.variables = c("rep", "site"), .parallel = T)
save(sst_ALL_KS, file = "data/sst_ALL_KS.Rdata")
rm(sst_ALL_clim_only, sst_ALL_KS); gc()load("data/sst_ALL_KS.Rdata")
# Filter non-significant results
KS_sig <- sst_ALL_KS %>%
filter(p.value <= 0.05) %>%
group_by(site, metric, year_2) %>%
summarise(count = n()) %>%
ungroup()
# Pad in NA for years with no significant differences
KS_sig <- padr::pad_int(x = KS_sig, by = "year_2", group = c("site", "metric")) %>%
select(site, metric, year_2, count) %>%
rbind(data.frame(site = "Med", metric = "seas", year_2 = 25:37, count = NA))
ggplot(KS_sig, aes(x = year_2, y = site)) +
geom_line(aes(colour = count), size = 3) +
geom_vline(aes(xintercept = 30), alpha = 0.7, linetype = "dotted") +
facet_wrap(~metric, ncol = 1) +
scale_colour_viridis_c() +
scale_x_reverse(breaks = seq(5, 35, 5)) +
labs(x = "Climatology period (years)",
y = NULL,
colour = "Sig. dif.\nout of 100")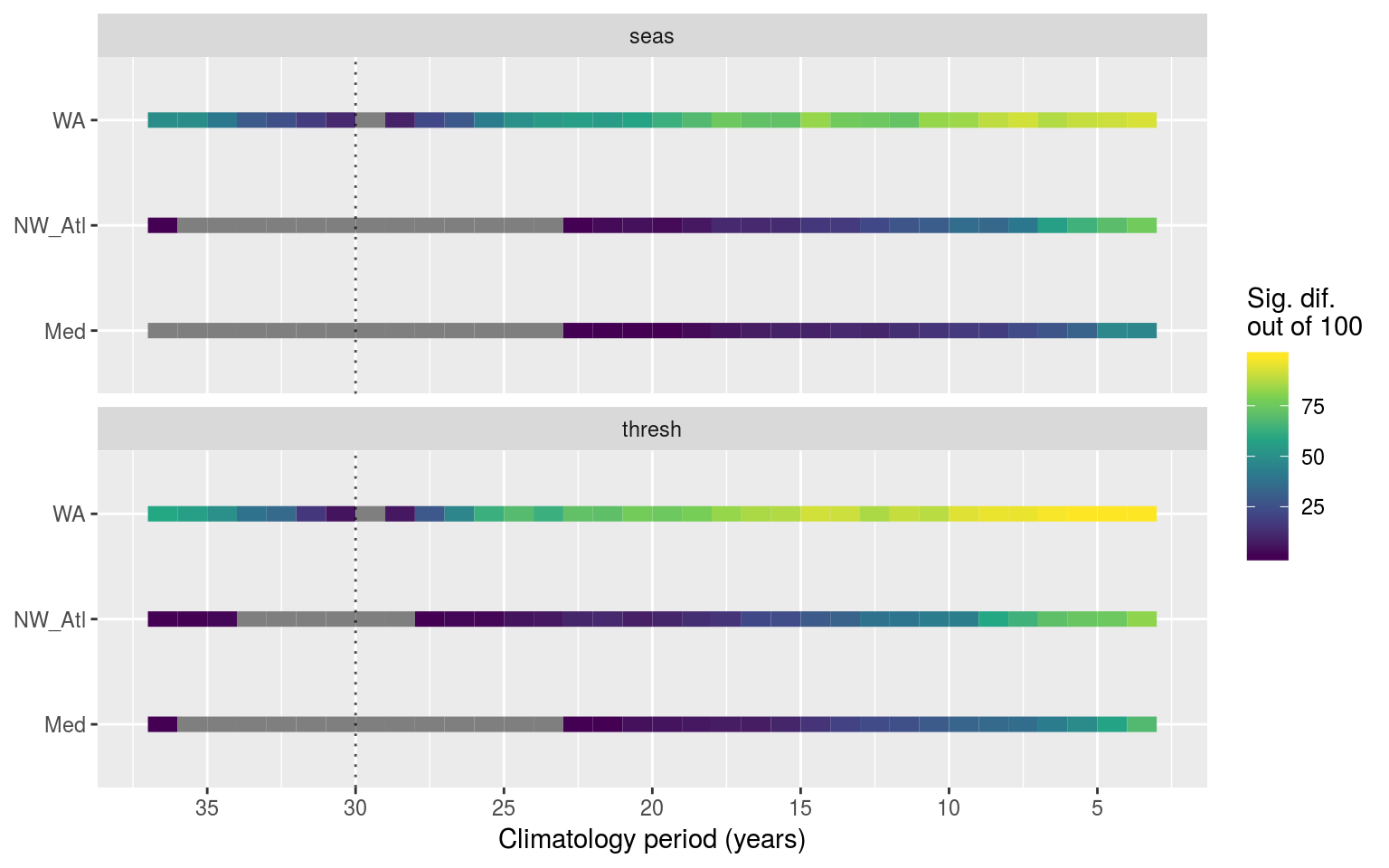
Line plots showing the results of pairwise Kolmogorov-Smirnoff tests for the seasonal signals (top panel) and 90th percentile thresholds (bottom panel) from the same time series at differing lengths. The colour of the line shows how many times out of 100 re-samples that the climatologies were significantly different from the control. The dotted verticle line denotes the 30 year climatology mark, against which all othe climatologies were compared. If no re-samplings were significantly different this is shown with a grey line.
Expand here to see past versions of KS-clims-1.png:
| Version | Author | Date |
|---|---|---|
| fa7fd57 | robwschlegel | 2019-03-19 |
rm(sst_ALL_KS, KS_sig); gc() used (Mb) gc trigger (Mb) max used (Mb)
Ncells 2102814 112.4 3944070 210.7 3944070 210.7
Vcells 3894847 29.8 10146329 77.5 10146143 77.5The above figure shows that the results of the KS tests were more often significantly different than the ANOVAs. This means that the actual shape of the distributions changes rather quickly, as shown by how it only takes a few years for the KS test to show the results as being different. The ANOVAs report fewer significant differences because that is a test of central tendency. Meaning that the shape of the climatologies changes rather quickly depending on the climatology period used, but the overall range of values within a climatology does not differ significantly as quickly.
load("data/sst_ALL_clim_only.Rdata")
sst_ALL_clim_1 <- sst_ALL_clim_only %>%
filter(rep == "1") %>%
arrange(year_index)
ggplot(data = sst_ALL_clim_1, aes(x = doy, y = thresh)) +
geom_line(aes(group = year_index, colour = year_index), alpha = 0.8) +
facet_wrap(~site, ncol = 1) +
scale_colour_viridis_c(direction = -1)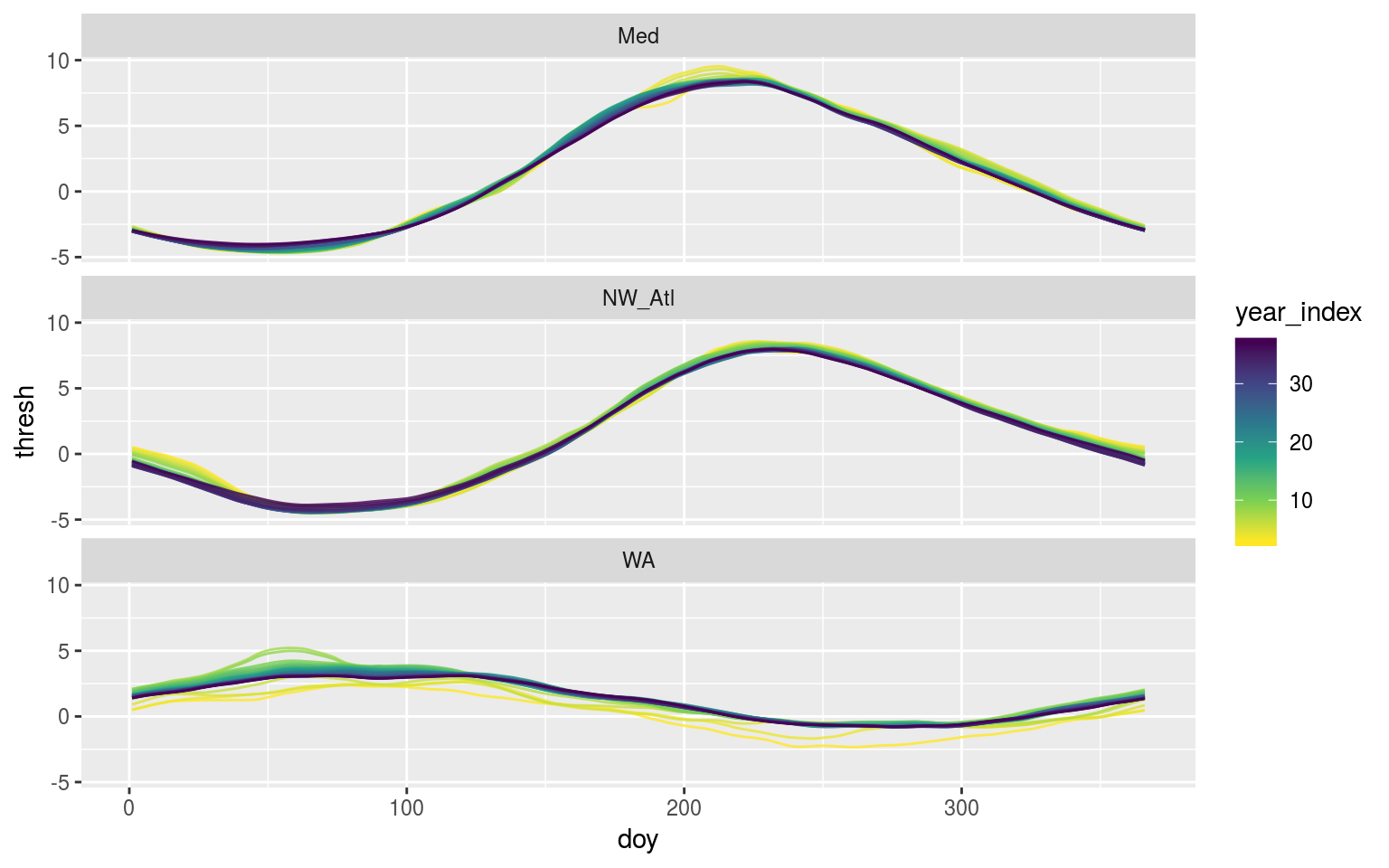
Time series of each of climatology period used from the original data shown overlapping one another to visualise how the climatologies differ depending on the length of the climatology period used.
Expand here to see past versions of clim-ts-all-1.png:
| Version | Author | Date |
|---|---|---|
| fa7fd57 | robwschlegel | 2019-03-19 |
The above figure shows us that the larger the seasonal signal is, the less effect deviations away from it will likely have.
Linear relationships (R^2)
It is also of interest to see what the relationship in the mean and SD of the seasonal signal and thresholds may be against the length of the time series.
load("data/sst_ALL_clim_only.Rdata")
# Summary stats for correlation
sst_ALL_smooth_R2_base <- sst_ALL_clim_only %>%
group_by(rep, site, year_index) %>%
summarise(seas_mean = mean(seas),
seas_sd = sd(seas),
thresh_mean = mean(thresh),
thresh_sd = sd(thresh)) %>%
# select(-year_index) %>%
group_by(rep, site)
save(sst_ALL_smooth_R2_base, file = "data/sst_ALL_smooth_R2_base.Rdata")
rm(sst_ALL_clim_only, sst_ALL_smooth_R2_base); gc()load("data/sst_ALL_smooth_R2_base.Rdata")
sst_ALL_smooth_R2_base_long <- sst_ALL_smooth_R2_base %>%
gather(key = metric, value = val, -year_index, -site, -rep)
ggplot(sst_ALL_smooth_R2_base_long, aes(x = year_index, y = val)) +
geom_point() +
geom_smooth(method = "lm") +
facet_grid(metric~site, scales = "free_y")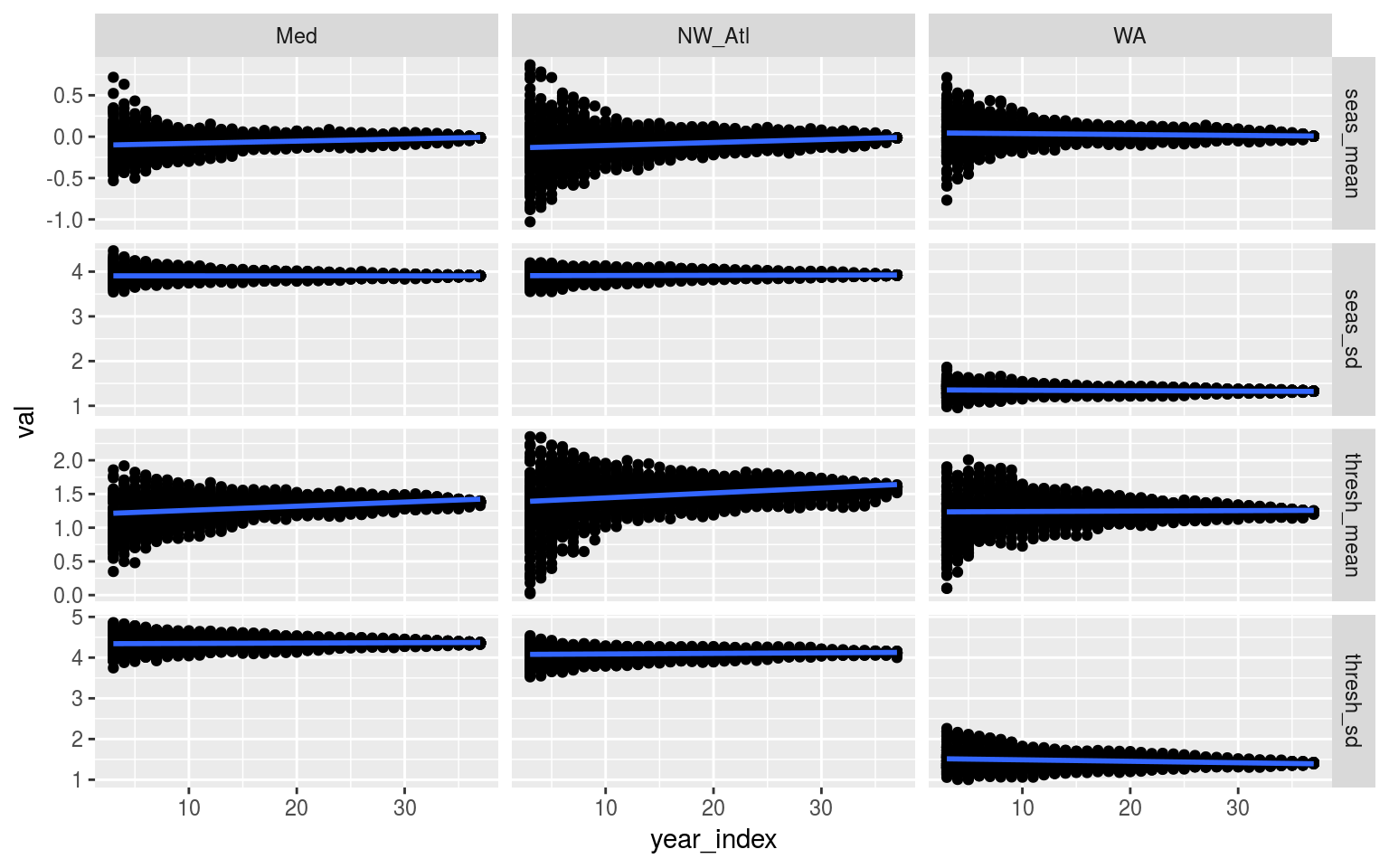
Scatterplot showing the relationship between time series length (x-axis) and a range of metrics (y-axis). The clear pattern is that the variance of the resulting relationships diminishes rapidly after at least 10 years of data are being used to determine the seasonal signal and threshold. The data shown here are all 100 re-samples.
Expand here to see past versions of clim-R2-base-scatter-plot-1.png:
| Version | Author | Date |
|---|---|---|
| fa7fd57 | robwschlegel | 2019-03-19 |
# Load base data
load("data/sst_ALL_smooth_R2_base.Rdata")
# Linear model function for nesting
linear_vals <- function(df, val){
val_vector <- as.vector(as.matrix(df[,colnames(df) == val]))
year_vector <- df$year_index
l_mod <- lm(val_vector ~ year_vector)
res <- glance(l_mod) %>%
mutate(metric = val,
adj.r.squared = round(adj.r.squared, 2),
p.value = round(p.value, 4)) %>%
select(metric, adj.r.squared, p.value)
row.names(res) <- NULL
return(res)
}
# Simple wrapper
linear_vals_wrap <- function(df){
seas_mean <- linear_vals(df, "seas_mean")
seas_sd <- linear_vals(df, "seas_sd")
thresh_mean <- linear_vals(df, "thresh_mean")
thresh_sd <- linear_vals(df, "thresh_sd")
res <- rbind(seas_mean, seas_sd, thresh_mean, thresh_sd)
return(res)
}
# Calculate all R2 and p values
sst_ALL_smooth_R2 <- sst_ALL_smooth_R2_base %>%
group_by(site, rep) %>%
nest() %>%
mutate(lm_res = map(data, linear_vals_wrap)) %>%
select(-data) %>%
unnest() %>%
mutate(adj.r.squared = ifelse(adj.r.squared < 0, 0, adj.r.squared))
# Save and clear
save(sst_ALL_smooth_R2, file = "data/sst_ALL_smooth_R2.Rdata")load("data/sst_ALL_smooth_R2.Rdata")
ggplot(sst_ALL_smooth_R2, aes(x = metric, y = adj.r.squared, fill = metric)) +
geom_boxplot() +
scale_fill_viridis_d() +
facet_wrap(~site)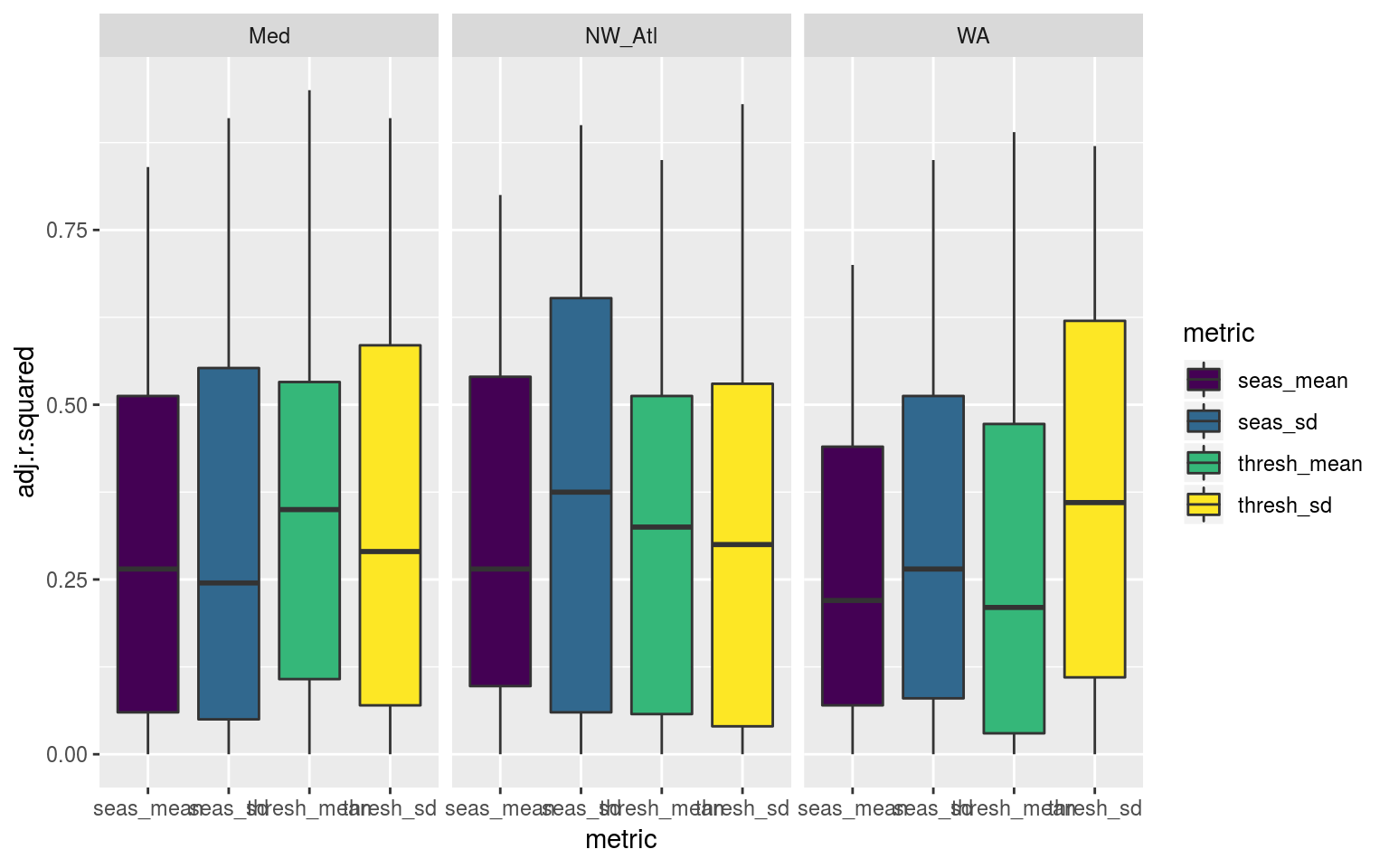
Boxplots showing the distribution of coefficients of determination (R2) for the linear relationship between the mean and SD of the climatologies for all the varying lengths of the 100 re-samples of each of the three reference time series.
Expand here to see past versions of clim-R2-fig-1.png:
| Version | Author | Date |
|---|---|---|
| fa7fd57 | robwschlegel | 2019-03-19 |
rm(sst_ALL_smooth_R2)MHW metrics
With the effects of time series duration on the climatologies now mapped out it is time to apply a similar methodology to the MHW metrics. In order to produce more robust statistics we will now use only results from time series of lengths 37 –10 years and will only compare events that occurred in the most recent 10 years of each time series.
# Quick wrapper for getting results for ANOVA on events
# df <- sst_ALL_smooth %>%
# filter(rep == "1", site == "WA") %>%
# select(-rep, -site)
event_aov_tukey <- function(df){
res <- df %>%
unnest(events) %>%
filter(row_number() %% 2 == 0) %>%
unnest(events) %>%
filter(year_index >= 10) %>%
filter(date_start >= "2009-01-02", date_end <= "2018-12-31") %>%
select(year_index, duration, intensity_mean, intensity_max, intensity_cumulative) %>%
mutate(year_index = as.factor(year_index)) %>%
nest() %>%
mutate(aov = map(data, aov_p),
tukey = map(data, aov_tukey)) %>%
select(-data)
return(res)
}ANOVA p-values
load("data/sst_ALL_smooth.Rdata")
doMC::registerDoMC(cores = 50)
sst_ALL_event_aov_tukey <- plyr::ddply(sst_ALL_smooth, c("rep", "site"),
event_aov_tukey, .parallel = T)
save(sst_ALL_event_aov_tukey, file = "data/sst_ALL_event_aov_tukey.Rdata")# This file is not uploaded to GitHub as it is too large
# One must first run the above code locally to generate and save the file
load("data/sst_ALL_event_aov_tukey.Rdata")
sst_ALL_event_aov <- sst_ALL_event_aov_tukey %>%
select(-tukey) %>%
unnest()
label_count_event_aov_p <- sst_ALL_event_aov %>%
group_by(site, metric) %>%
filter(p.value <= 0.05) %>%
summarise(count = n())
knitr::kable(label_count_event_aov_p,
col.names = c("Time series", "MHW metric", "Significantly different"),
caption = "Table 1: This table shows the results of 100 ANOVAs on event metrics detected in the real and re-sampled time series with systematically shortened lengths from 37 -- 10 years.")| Time series | MHW metric | Significantly different |
|---|---|---|
| NW_Atl | duration | 1 |
| NW_Atl | intensity_mean | 2 |
| WA | intensity_max | 1 |
| WA | intensity_mean | 5 |
The above table shows us that generally the length of a time series has no significant effect on the MHWs detected within it. We see that the MHWs in the Mediterranean time series are not affected at all in any of the 100 replicates. Surprisingly the events in the North West Atlantic time series are significantly different more often than the Western Australia time series. This must mean that the event metrics are more sensistive to a number of longer larger events than a single large one, as is the case in the WA time series. This also means that the thresholds between shortened time series may not need to be significantly different in order to produce significantly different events. It must now be investigate just which years are significantly different from one another.
Post-hoc Tukey test
sst_ALL_event_tukey <- sst_ALL_event_aov_tukey %>%
select(-aov) %>%
unnest()
event_tukey_sig <- sst_ALL_event_tukey %>%
filter(adj.p.value <= 0.05)# %>%
# separate(comparison, into = c("year_long", "year_short"), sep = "-") %>%
# mutate(year_long = as.numeric(year_long),
# year_short = as.numeric(year_short)) %>%
# mutate(year_dif = year_long - year_short) %>%
# # select(-adj.p.value, -rep) %>%
# group_by(site, metric, year_long, year_short, year_dif) %>%
# summarise(count = n()) %>%
# ungroup() %>%
# arrange(-year_long, -year_dif) %>%
# mutate(year_index = factor(paste0(year_long,"-",year_dif)),
# year_index = factor(year_index, levels = unique(year_index)))
# ggplot(event_tukey_sig, aes(x = year_long, y = year_short)) +
# # geom_point() +
# # geom_errorbarh(aes(xmin = year_long, xmax = year_short)) +
# geom_tile(aes(fill = count)) +
# facet_grid(~metric) +
# scale_x_reverse(breaks = seq(3, 36, 3)) +
# scale_y_reverse(breaks = seq(3 , 7, 1)) +
# scale_fill_viridis_c("count out of \n100 re-samples", breaks = seq(2,12,2)) +
# labs(x = "Length (years) of long time series",
# y = "Length (years) of short time series")The post-hoc Tukey test revealed that only 1 rep out of 100 for the WA produced a significant difference when comparing the 10 year time series against 28, 29, 36, and 37 year time series. This shows that time series length has a negligible effect on the detection of events.
Linear relationships (R^2)
load("data/sst_ALL_smooth.Rdata")
sst_ALL_event_cor <- sst_ALL_smooth %>%
unnest(events) %>%
filter(row_number() %% 2 == 0) %>%
unnest(events) %>%
filter(year_index >= 10) %>%
filter(date_start >= "2009-01-01", date_end <= "2018-12-31") %>%
select(rep, site, year_index, duration, intensity_mean, intensity_max, intensity_cumulative) %>%
group_by(site, rep, year_index) %>%
summarise_all(.funs = c("mean", "sd")) %>%
group_by(site, rep) %>%
do(data.frame(t(cor(.[,4:11], .[,3])))) %>%
gather(key = y, value = r, -rep, -site) %>%
mutate(x = "length")
save(sst_ALL_event_cor, file = "data/sst_ALL_event_cor.Rdata")load("data/sst_ALL_event_cor.Rdata")
ggplot(sst_ALL_event_cor, aes(x = y, y = r, fill = y)) +
geom_boxplot() +
geom_hline(aes(yintercept = 0), colour = "grey20",
size = 1.2, linetype = "dashed") +
scale_fill_viridis_d() +
facet_wrap(~site, ncol = 1) +
labs(x = NULL, y = "Pearson r") +
theme(axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
legend.position = "bottom")Confidence intervals
After looking at these correlations it seems as though somehow calculating confidence intervals on something would be worthwhile…
# Calculate CIs using a bootstrapping technique to deal with the non-normal small sample sizes
# df <- sst_ALL_both_event %>%
# filter(lubridate::year(date_peak) >= 2012,
# site == "WA",
# trend == "detrended") %>%
# select(year_index, duration, intensity_mean, intensity_max, intensity_cumulative) #%>%
# select(site, trend, year_index, duration, intensity_mean, intensity_max, intensity_cumulative) #%>%
# nest(-site, -trend)
Bmean <- function(data, indices) {
d <- data[indices] # allows boot to select sample
return(mean(d))
}
event_aov_CI <- function(df){
df_conf <- gather(df, key = "metric", value = "value", -year_index) %>%
group_by(year_index, metric) %>%
# ggplot(aes(x = value)) +
# geom_histogram(aes(fill = metric)) +
# facet_wrap(~metric, scales = "free_x")
summarise(lower = boot.ci(boot(data=value, statistic=Bmean, R=1000),
type = "basic")$basic[4],
mid = mean(value),
upper = boot.ci(boot(data=value, statistic=Bmean, R=1000),
type = "basic")$basic[5],
n = n()) %>%
mutate_if(is.numeric, round, 4)
return(df_conf)
}Categories
In this section we want to look at how the categories in the different time periods compare. I’ll start out with doing basic calculations and comparisons of the categories of events with the different time periods. After that we will run chi-squared tests. Lastly a post-hoc test of sorts will be used to pull out exactly which categories differ significantly in count with different time series lengths.
Event count
load("data/sst_ALL_smooth.Rdata")
suppressWarnings(
sst_ALL_category <- sst_ALL_smooth %>%
select(-events) %>%
unnest(cats) %>%
filter(year_index >= 10,
peak_date >= "2009-01-01", peak_date <= "2018-12-31")
)
# Indeed... but how to compare qualitative data?
# Let's start out by counting the occurrence of the categories
sst_ALL_clim_category_count <- sst_ALL_category %>%
group_by(site, rep, year_index, category) %>%
summarise(count = n()) %>%
mutate(category = factor(category,
levels = c("I Moderate", "II Strong",
"III Severe", "IV Extreme")))
save(sst_ALL_clim_category_count, file = "data/sst_ALL_clim_category_count.Rdata")load("data/sst_ALL_clim_category_count.Rdata")
# total_category_count <- sst_ALL_clim_category_count %>%
# ungroup() %>%
# select(-rep) %>%
# group_by(site, category) %>%
# summarise(count = sum(count))
ggplot(sst_ALL_clim_category_count, aes(x = year_index, y = count, fill = category)) +
geom_bar(stat = "identity") +
# geom_label(data = label_category_count, show.legend = F, fill = "white",
# aes(label = count, y = 1000)) +
scale_fill_manual(values = MHW_colours) +
facet_wrap(~site, ncol = 1) +
labs(x = "Tim series length (years)", y = "Total count") +
theme(legend.position = "bottom")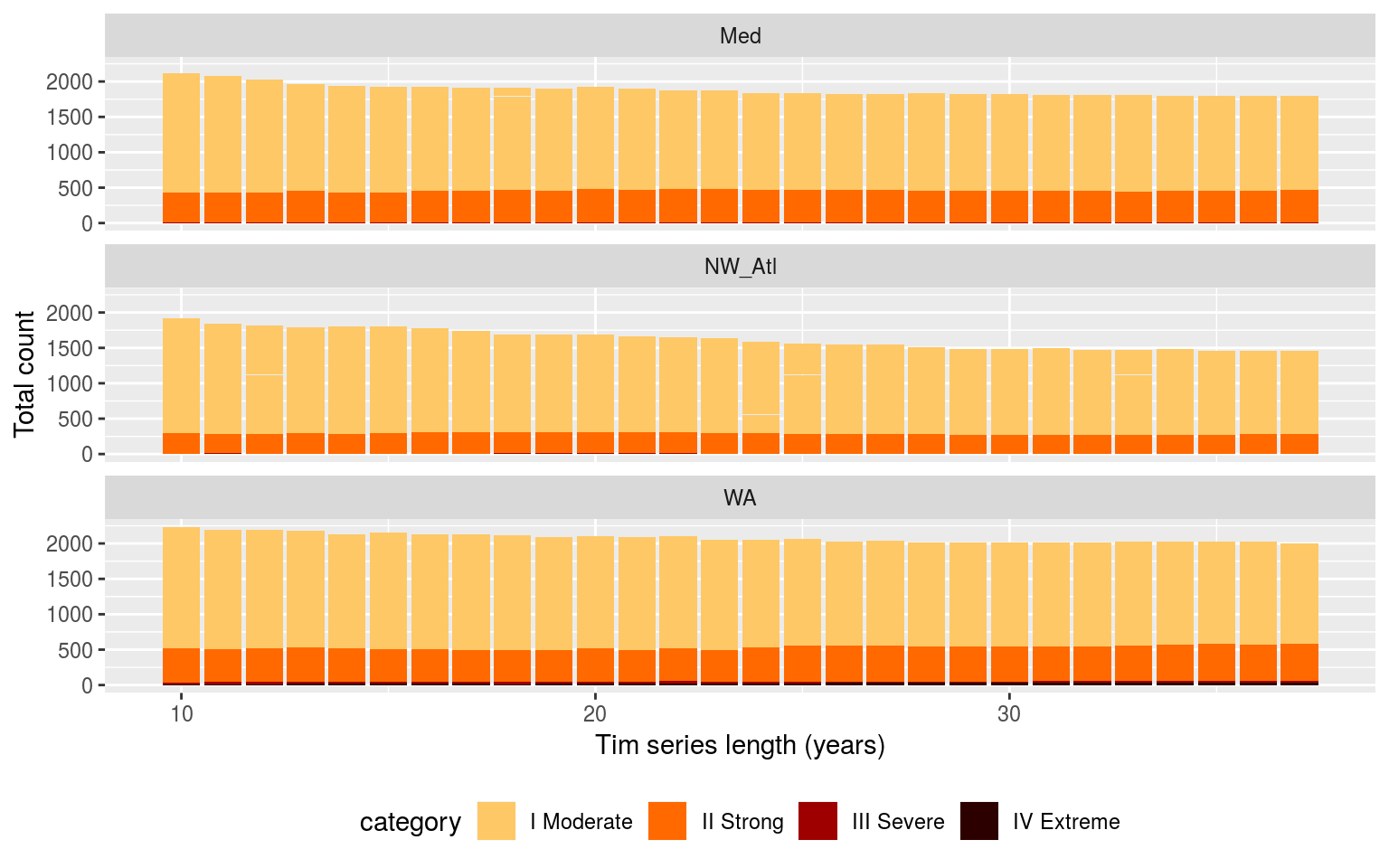
Bar plots showing the counts of the different categories of events faceted in a grid with different sites along the top and the different category classifications down the side. The colours of the bars denote the different climatology periods used. Note that the general trend is that more ‘smaller’ events are detected with a 10 year clim period, and more ‘larger’ events detected with the 30 year clim. The 20 year clim tends to rest in the middle.
Expand here to see past versions of cat-compare-1.png:
| Version | Author | Date |
|---|---|---|
| fa7fd57 | robwschlegel | 2019-03-19 |
The results in the figure above show that we see more larger events when the climatologies are based on 30 years of data, rather than 10, but that we tend to see more events overall when we use only 10 years of data. This is about what would be expected if there were a trend present in the data. SO it is interesting that it still comes through.
chi-squared
In this section we will use pairwise chi-squared tests to see if the count of events in the different re-sampled lengths differ significantly from the same re-sampled time series of 30 years (this serves as the control). We will combine categories 3 and 4 together into one category for a more robust test and because we are interested in how these larger events stand out together against the smaller events.
# Load and extract category data
load("data/sst_ALL_smooth.Rdata")
suppressWarnings( # Suppress warning about category levels not all being present
sst_ALL_cat <- sst_ALL_smooth %>%
select(-events) %>%
unnest()
)
# Prep the data for better chi-squared use
sst_ALL_cat_prep <- sst_ALL_cat %>%
filter(year_index >= 10,
peak_date >= "2009-01-01", peak_date <= "2018-12-31") %>%
select(year_index, rep, site, category) %>%
mutate(category = ifelse(category == "III Severe", "III & IV", category),
category = ifelse(category == "IV Extreme", "III & IV", category))
# Extract the true climatologies
sst_ALL_cat_only_30 <- sst_ALL_cat_prep %>%
filter(year_index == 30) %>%
select(site, year_index, rep, category)
# chi-squared pairwise function
# Takes one rep of missing data and compares it against the complete data
# df <- sst_ALL_miss_cat_prep %>%
# filter(site == "WA", rep == "20", miss == 0.1)
# df <- unnest(slice(sst_ALL_miss_cat_chi,1)) %>%
# select(-site2, -rep, -miss2)
chi_pair <- function(df){
# Prep data
df_comp <- sst_ALL_cat_only_30 %>%
filter(site == df$site[1],
rep == df$rep[1])
df_joint <- rbind(df, df_comp)
# Run tests
res <- chisq.test(table(df_joint$year_index, df_joint$category), correct = FALSE)
res_broom <- broom::augment(res) %>%
mutate(p.value = broom::tidy(res)$p.value)
return(res_broom)
}
# The pairwise chai-squared results
suppressWarnings( # Suppress poor match warnings
sst_ALL_cat_chi <- sst_ALL_cat_prep %>%
filter(year_index != 30) %>%
# mutate(miss = as.factor(miss)) %>%
mutate(site2 = site,
year_index2 = year_index,
rep2 = rep) %>%
group_by(site2, year_index2, rep2) %>%
nest() %>%
mutate(chi = map(data, chi_pair)) %>%
select(-data) %>%
unnest() %>%
dplyr::rename(site = site2, year_index = year_index2, rep = rep2,
year_comp = Var1, category = Var2)
)
save(sst_ALL_cat_chi, file = "data/sst_ALL_cat_chi.Rdata")load("data/sst_ALL_cat_chi.Rdata")
sst_ALL_cat_chi_sig <- sst_ALL_cat_chi %>%
filter(p.value <= 0.05) %>%
select(site, year_index, p.value) %>%
group_by(site, year_index) %>%
unique() %>%
summarise(count = n())
knitr::kable(sst_ALL_cat_chi_sig, caption = "Table showing the count out of 100 for significant differences (_p_-value) for the count of different categories of events for each site based on the amount of missing data present.")load("data/sst_ALL_cat_chi.Rdata")
# Prep data for plotting
chi_sig <- sst_ALL_cat_chi %>%
filter(p.value <= 0.05) %>%
select(site, year_index, p.value) %>%
group_by(site, year_index) %>%
unique() %>%
summarise(count = n())
ggplot(chi_sig, aes(x = as.numeric(year_index), y = site)) +
geom_line(aes(colour = count, group = site)) +
geom_point(aes(colour = count)) +
# facet_wrap(~metric, ncol = 1) +
scale_colour_viridis_c() +
scale_x_reverse() +
labs(x = "Time series length (years)", y = NULL,
colour = "Sig. dif.\nout of 100") +
theme(axis.text.x = element_text(angle = 20))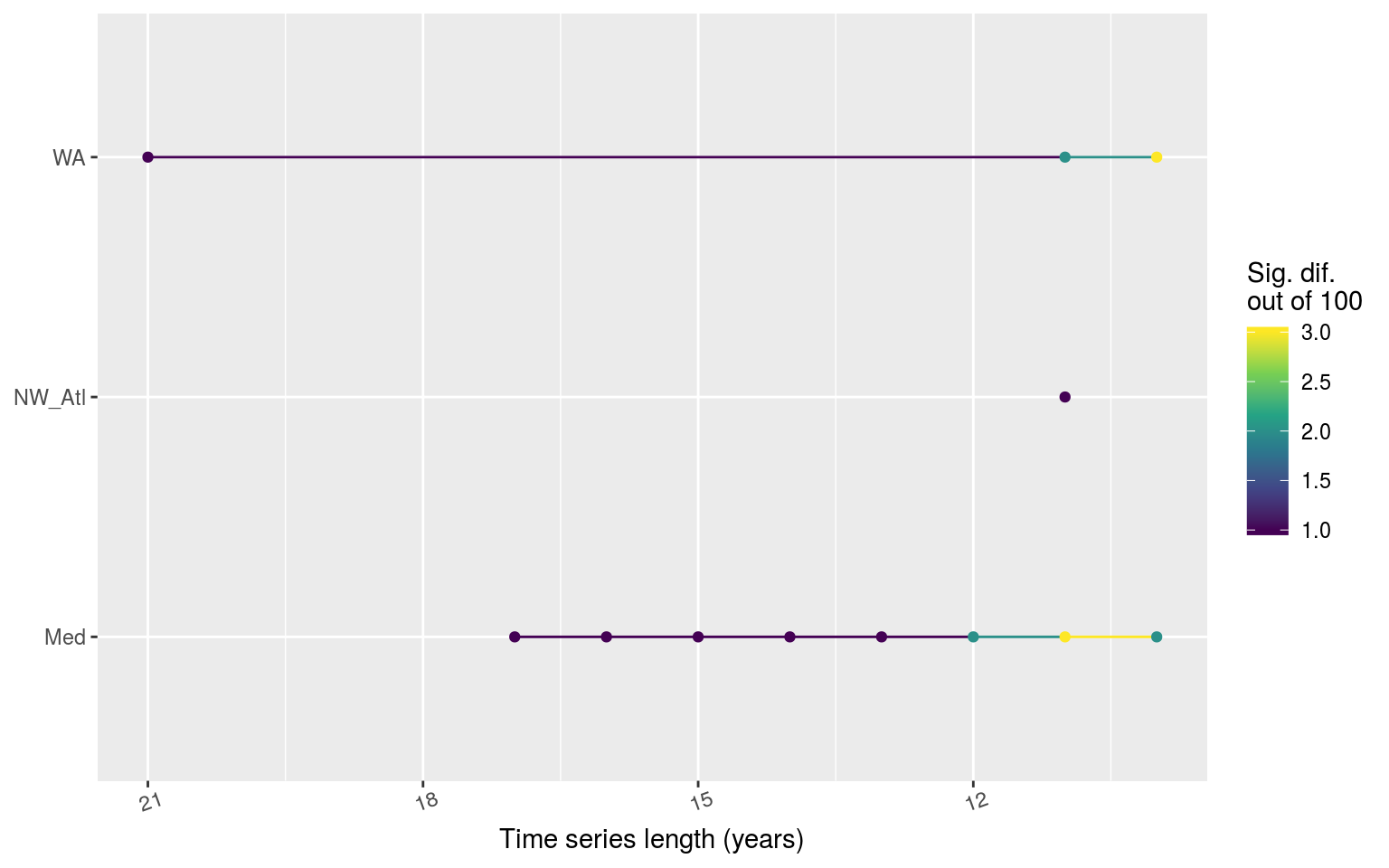
Line graph showing the count of times out of 100 random replicates when a given time series length led to significant differences in the count of categories of MHWs as determined by a chi-squared test.
Expand here to see past versions of chi-pair-plot-1.png:
| Version | Author | Date |
|---|---|---|
| fa7fd57 | robwschlegel | 2019-03-19 |
The line plot above shows at what point the category counts begin to differ significantly from the matching 30 year time series. It was expected that the WA time series would show sig. dif. more quickly than the other two, though I had expected the Med to be the slowest to change, not the NW_Atl. Over a sample of 100, there were never more than five replicates showing significant differences in count of categories though so it is relatively safe to say that the length of a time series has little effect on the count of events.
Post-hoc chi-squared
It is also necessary to see which of the categories specifically are differing significantly. Unfortunately there is no proper post-hoc chi-squared test. Instead it is possible to look at the standardised residuals for each category within the pairwise comparisons being made.
I still need to do this…
Correlations
It would also be useful to look at the correlations between consecutive missing days of data and significantly different counts of categories.
Can we fix it?
Fourier transform climatologies
Lastly we will now go about reproducing all of the checks made above, but based on a climatology derived from a Fourier transformation, and not the default climatology creation method.
(RWS: I’ve not gotten to the Fourier transformations yet either. I’m also not convinced that this is going to be the way forward. I am thinking that it may be better to just increase the smoothing window width to produce a more sinusoidal climatology line. But I suppose that in order to know that it must be tested.)
(ECJO)[One advantage of the Fourier transform method (or, I think equivalently harmonic regression) is that it is VERY fast. Much faster than the method used in the MHW code. I also think you can probably get away with shorter time series using this method than with the “standard MHW climatology” method]
(RWS)[I suppose then that I will need to look into this.]
Wider windows
Talk a bit here about changing the arguments for climatology creation to get them closer to the control.
Best practices
(RWS: I envision this last section distilling everything learned above into a nice bullet list. These bulletted items for each different vignette would then all be added up in one central best practices vignette that would be ported over into the final write-up in the discussion/best-practices section of the paper. Ideally these best-practices could also be incorporated into the R/python distributions of the MHW detection code to allow users to make use of the findings from the paper in their own work.)
Options for improving climatology precision in short time series
Options for improving MHW metric precision in short time series
References
Oliver, Eric C.J., Markus G. Donat, Michael T. Burrows, Pippa J. Moore, Dan A. Smale, Lisa V. Alexander, Jessica A. Benthuysen, et al. 2018. “Longer and more frequent marine heatwaves over the past century.” Nature Communications 9 (1). doi:10.1038/s41467-018-03732-9.
Schlegel, Robert W., and Albertus J. Smit. 2016. “Climate Change in Coastal Waters: Time Series Properties Affecting Trend Estimation.” Journal of Climate 29 (24): 9113–24. doi:10.1175/JCLI-D-16-0014.1.
Session information
sessionInfo()R version 3.5.3 (2019-03-11)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 16.04.5 LTS
Matrix products: default
BLAS: /usr/lib/openblas-base/libblas.so.3
LAPACK: /usr/lib/libopenblasp-r0.2.18.so
locale:
[1] LC_CTYPE=en_CA.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_CA.UTF-8 LC_COLLATE=en_CA.UTF-8
[5] LC_MONETARY=en_CA.UTF-8 LC_MESSAGES=en_CA.UTF-8
[7] LC_PAPER=en_CA.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_CA.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] parallel stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] bindrcpp_0.2.2 doMC_1.3.5 iterators_1.0.10
[4] foreach_1.4.4 mgcv_1.8-24 nlme_3.1-137
[7] FNN_1.1.2.1 boot_1.3-20 ggpubr_0.1.8
[10] magrittr_1.5 fasttime_1.0-2 lubridate_1.7.4
[13] heatwaveR_0.3.6.9003 data.table_1.11.6 broom_0.5.0
[16] forcats_0.3.0 stringr_1.3.1 dplyr_0.7.6
[19] purrr_0.2.5 readr_1.1.1 tidyr_0.8.1
[22] tibble_1.4.2 ggplot2_3.0.0 tidyverse_1.2.1
loaded via a namespace (and not attached):
[1] Rcpp_0.12.18 lattice_0.20-35 assertthat_0.2.0
[4] rprojroot_1.3-2 digest_0.6.16 R6_2.2.2
[7] cellranger_1.1.0 plyr_1.8.4 backports_1.1.2
[10] evaluate_0.11 highr_0.7 httr_1.3.1
[13] pillar_1.3.0 rlang_0.2.2 lazyeval_0.2.1
[16] readxl_1.1.0 rstudioapi_0.7 whisker_0.3-2
[19] R.utils_2.7.0 R.oo_1.22.0 Matrix_1.2-14
[22] rmarkdown_1.10 labeling_0.3 htmlwidgets_1.2
[25] munsell_0.5.0 compiler_3.5.3 modelr_0.1.2
[28] pkgconfig_2.0.2 htmltools_0.3.6 tidyselect_0.2.4
[31] workflowr_1.1.1 codetools_0.2-15 viridisLite_0.3.0
[34] crayon_1.3.4 withr_2.1.2 R.methodsS3_1.7.1
[37] grid_3.5.3 jsonlite_1.5 gtable_0.2.0
[40] git2r_0.23.0 scales_1.0.0 cli_1.0.0
[43] stringi_1.2.4 reshape2_1.4.3 padr_0.4.1
[46] xml2_1.2.0 tools_3.5.3 glue_1.3.0
[49] hms_0.4.2 yaml_2.2.0 colorspace_1.3-2
[52] rvest_0.3.2 plotly_4.8.0 knitr_1.20
[55] bindr_0.1.1 haven_1.1.2 This reproducible R Markdown analysis was created with workflowr 1.1.1