Brain Tumor-Flash
Yuxin Zou
2018-2-20
Last updated: 2018-02-27
Code version: 5769f40
R.matlab v3.6.1 (2016-10-19) successfully loaded. See ?R.matlab for help.
Attaching package: 'R.matlab'The following objects are masked from 'package:base':
getOption, isOpenBrain Tumor data:
The data consist of micro array gene expression measurements on 43 brain tumors of four types, together with 356 additional variables related to the expression data. We pre-processed the data by centering the data by column.
data = readMat('../data/BrainTumor.mat')
data = data$YFlash:
flash.data = flash_set_data(data)
f = flash(flash.data, greedy = TRUE, backfit = TRUE)
saveRDS(f, '../output/BrainTumorVarCol.rds')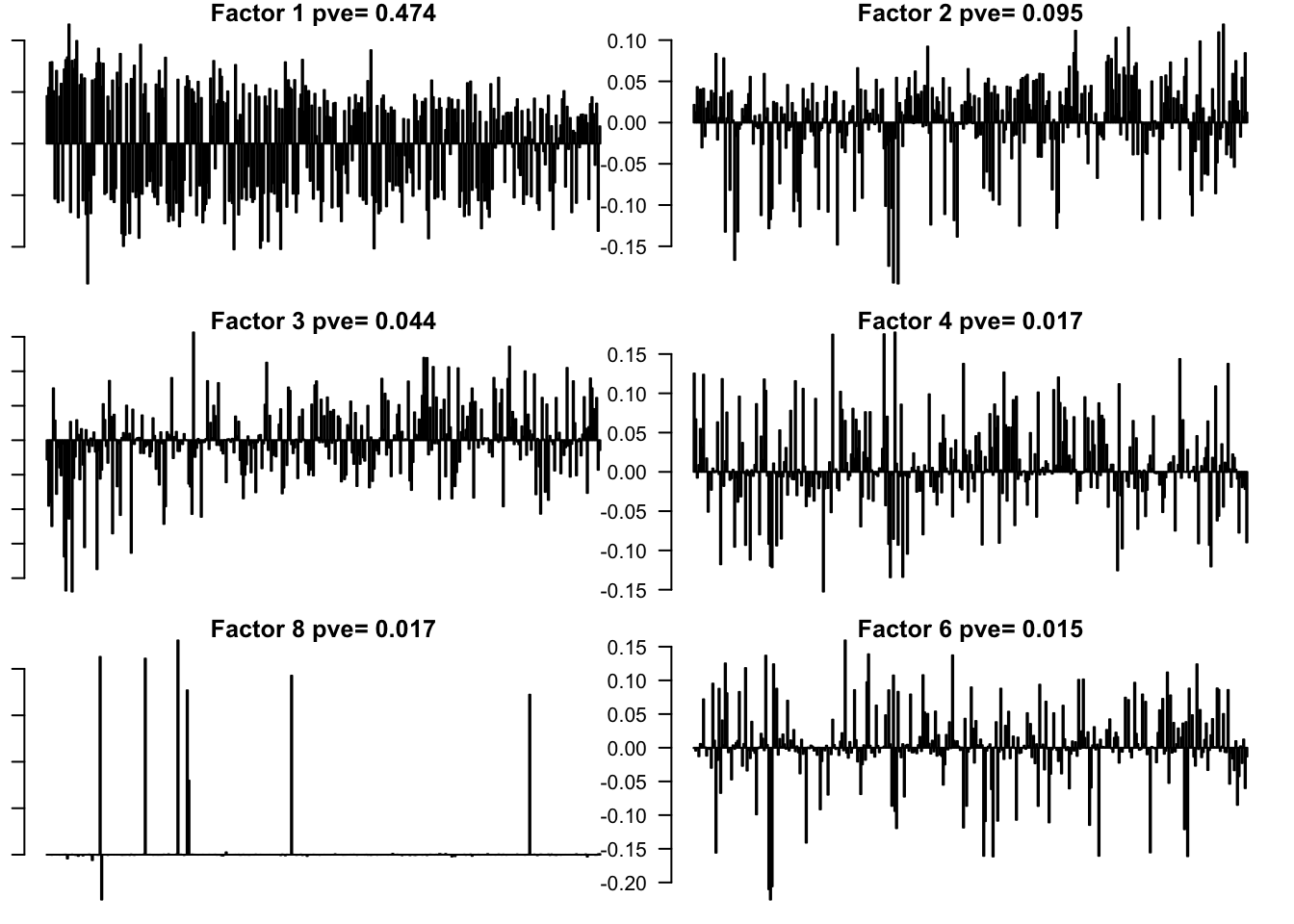
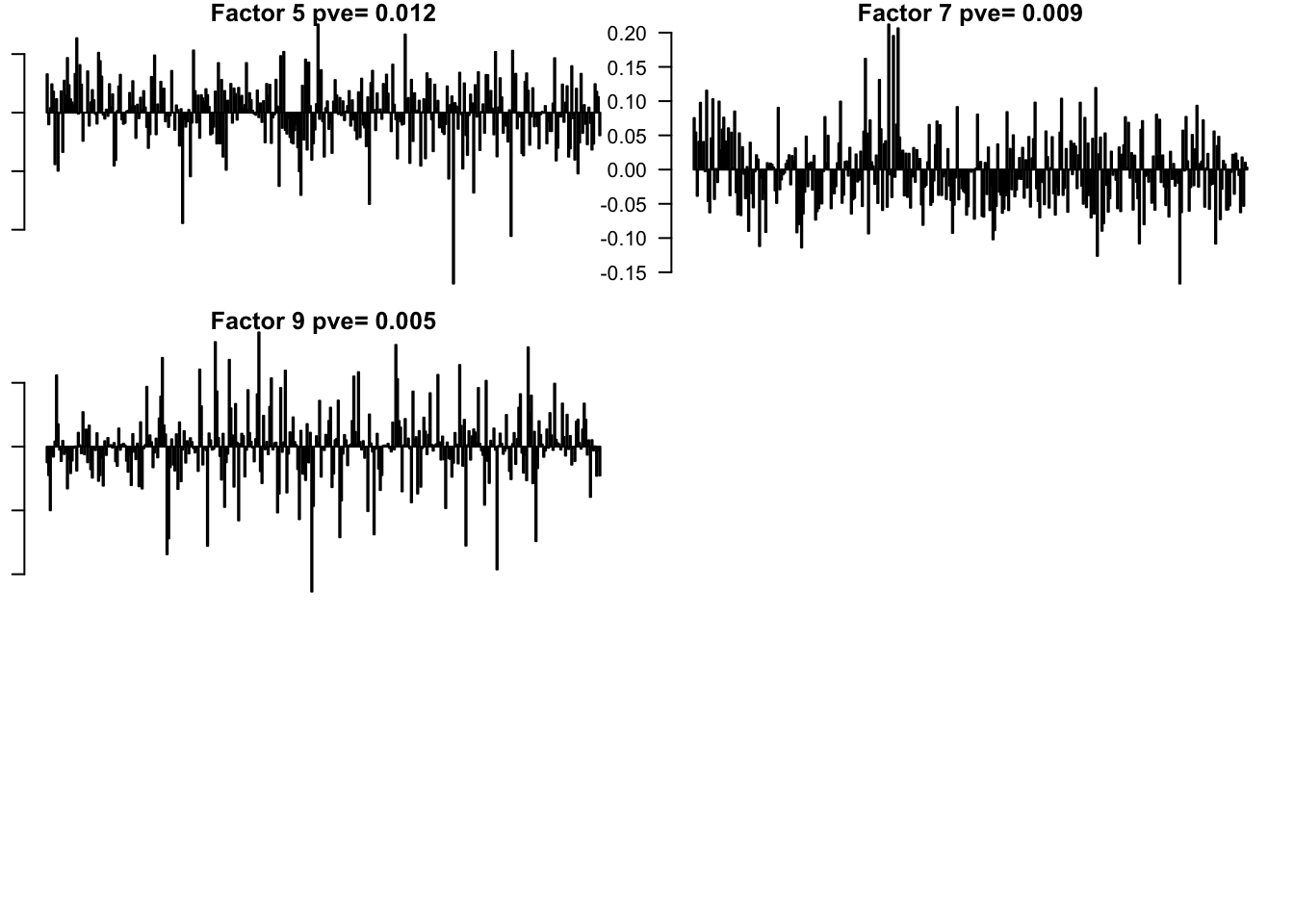
Flash again on the loading matrix
flash.loading = flash_set_data(fmodel$EL[,1:9])
flmodel = flash(flash.loading, greedy = TRUE, backfit = TRUE)The flash prefers the rank 0 model. There is no hidden structure in the loading matrix.
Session information
sessionInfo()R version 3.4.3 (2017-11-30)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS High Sierra 10.13.3
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] R.matlab_3.6.1 flashr_0.5-1
loaded via a namespace (and not attached):
[1] Rcpp_0.12.15 digest_0.6.13 rprojroot_1.2
[4] R.methodsS3_1.7.1 backports_1.1.2 git2r_0.20.0
[7] magrittr_1.5 evaluate_0.10.1 stringi_1.1.6
[10] R.utils_2.6.0 R.oo_1.21.0 rmarkdown_1.8
[13] tools_3.4.3 stringr_1.2.0 yaml_2.1.16
[16] compiler_3.4.3 htmltools_0.3.6 knitr_1.17 This R Markdown site was created with workflowr