Mash Missing Whole Row
Yuxin Zou
2018-2-28
Last updated: 2018-03-02
Code version: a79b566
R=5
library(mashr)Loading required package: ashrset.seed(2018)
data = simple_sims(500, err_sd = 0.1)
data.miss = data
missing.row = sample(1:nrow(data$Bhat), nrow(data$Bhat)/4)
missing.ind = matrix(0, nrow = nrow(data$B), ncol=ncol(data$B))
missing.ind[missing.row,] = 1
missing = which(missing.ind == 1)
data.miss$Bhat[missing] = NA
data.miss$Shat[missing] = NA
missing1 = is.na(data.miss$Bhat[,1])
# Set the missing value with large standard deviation
data.large.dev = data.miss
data.large.dev$Bhat[is.na(data.miss$Bhat)] = 0
data.large.dev$Shat[is.na(data.miss$Shat)] = 1000EE model
Warning in REBayes::KWDual(A, rep(1, k), normalize(w), control = control): estimated mixing distribution has some negative values:
consider reducing rtolWarning in mixIP(matrix_lik = structure(c(1, 0.204132576568413,
0.736530341806702, : Optimization step yields mixture weights that are
either too small, or negative; weights have been corrected and renormalized
after the optimization.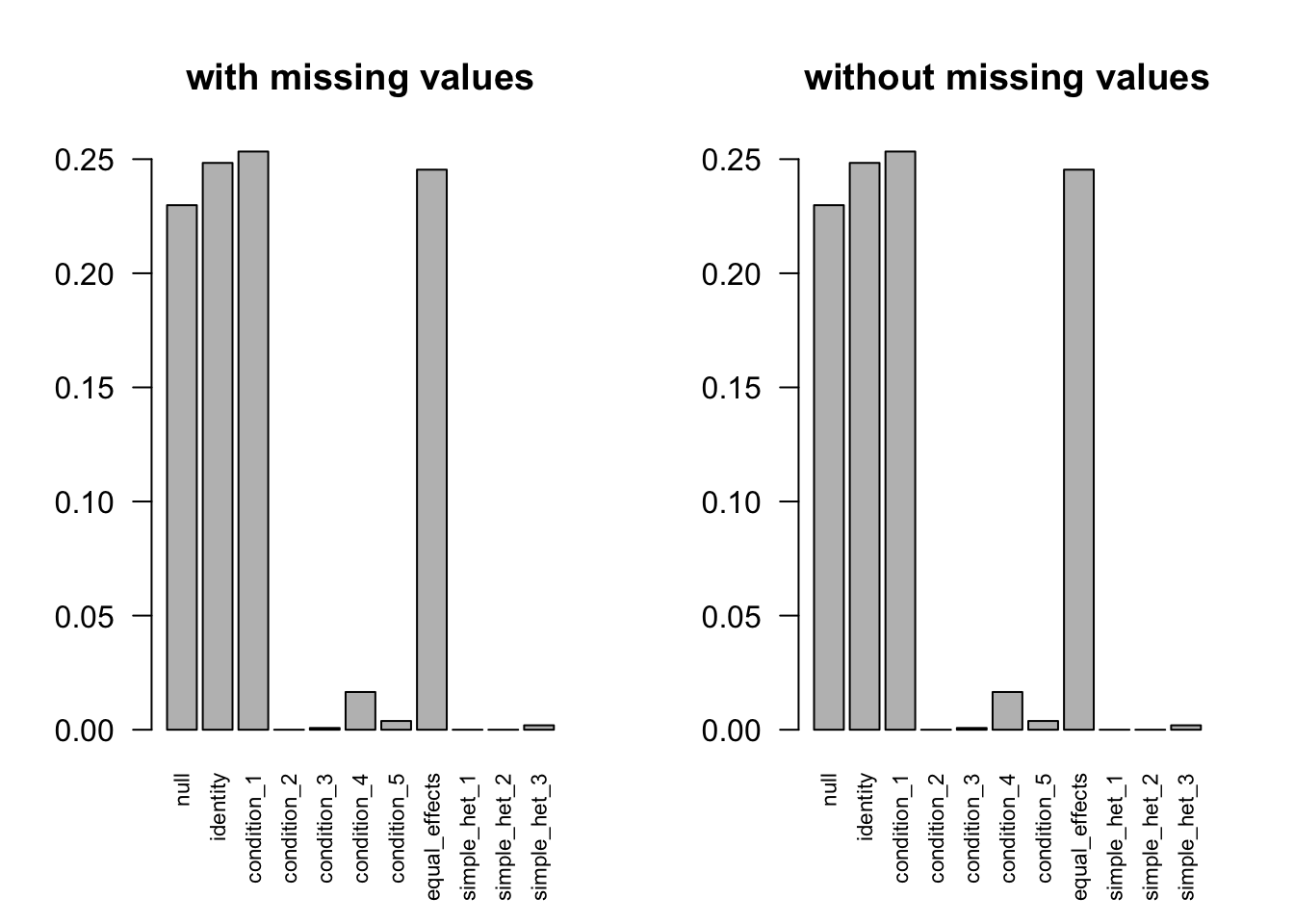
EZ model
FIXME: 'compute_posterior_matrices' in Rcpp does not transfer EZ to EEWarning in REBayes::KWDual(A, rep(1, k), normalize(w), control = control): estimated mixing distribution has some negative values:
consider reducing rtolWarning in mixIP(matrix_lik = structure(c(1, 0.204132576568412,
0.736530341806701, : Optimization step yields mixture weights that are
either too small, or negative; weights have been corrected and renormalized
after the optimization.FIXME: 'compute_posterior_matrices' in Rcpp does not transfer EZ to EE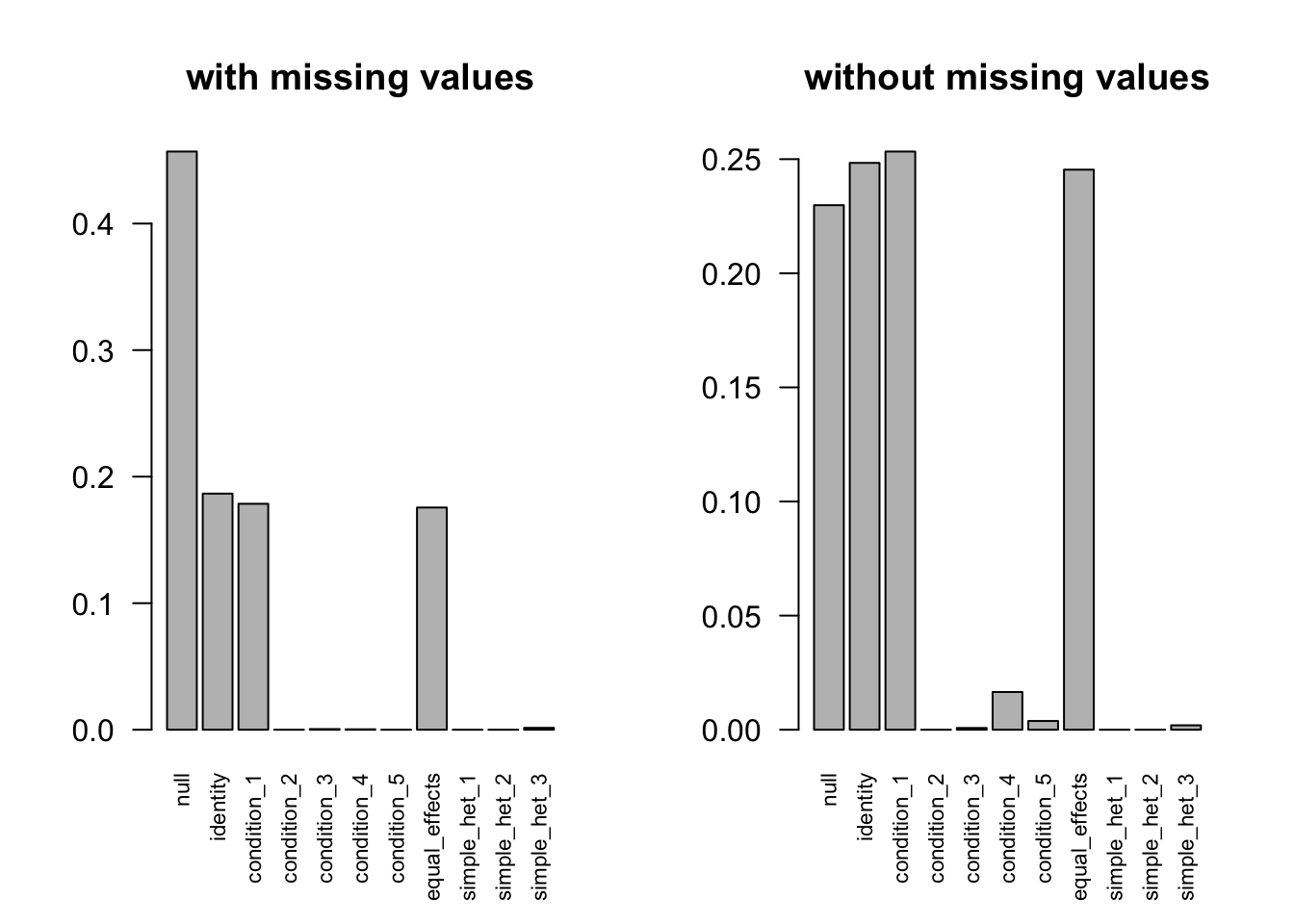
R=60
EE model
With missing values
 The weights learned from the data are correct.
The weights learned from the data are correct.
Without missing values
Warning in REBayes::KWDual(A, rep(1, k), normalize(w), control = control): estimated mixing distribution has some negative values:
consider reducing rtolWarning in mixIP(matrix_lik = structure(c(0.240920767436896,
0.337563330121546, : Optimization step yields mixture weights that are
either too small, or negative; weights have been corrected and renormalized
after the optimization.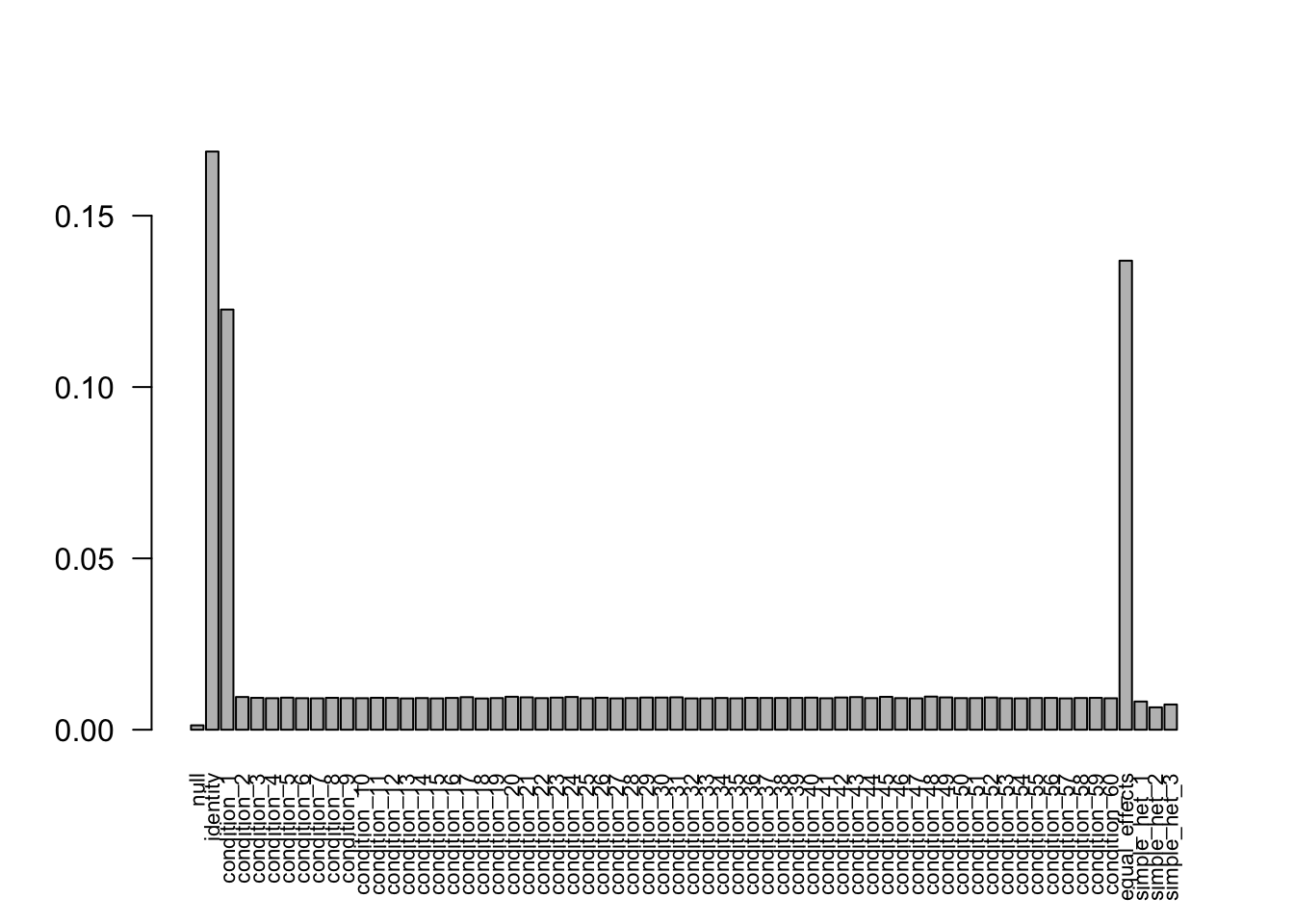 The estimated weights are very weird and incorrect. It ignores the null matrix.
The estimated weights are very weird and incorrect. It ignores the null matrix.
EZ model
With missing values
Warning in REBayes::KWDual(A, rep(1, k), normalize(w), control = control): estimated mixing distribution has some negative values:
consider reducing rtolWarning in mixIP(matrix_lik = structure(c(0.240920767436896,
0.33756333012155, : Optimization step yields mixture weights that are
either too small, or negative; weights have been corrected and renormalized
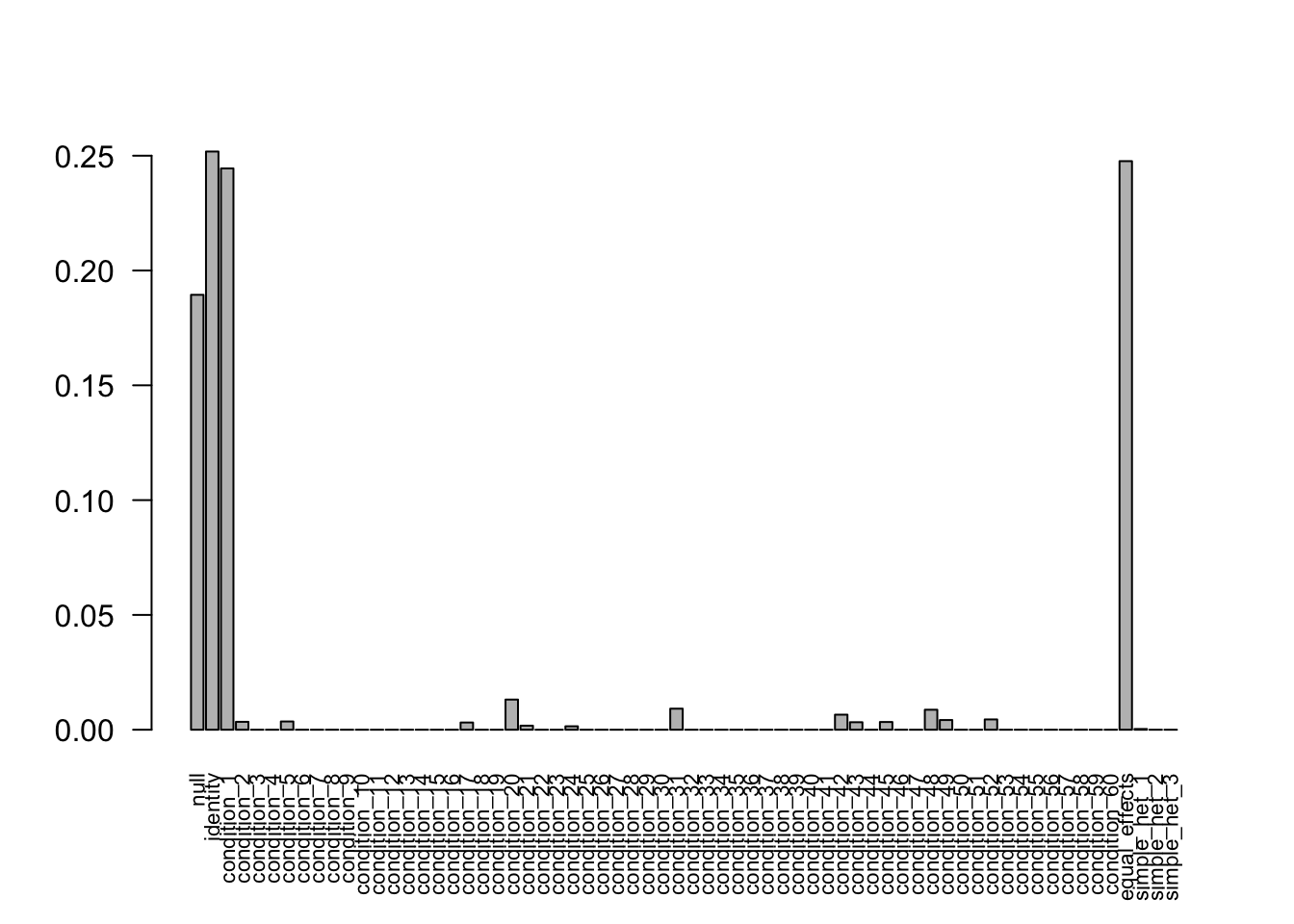
after the optimization.FIXME: 'compute_posterior_matrices' in Rcpp does not transfer EZ to EE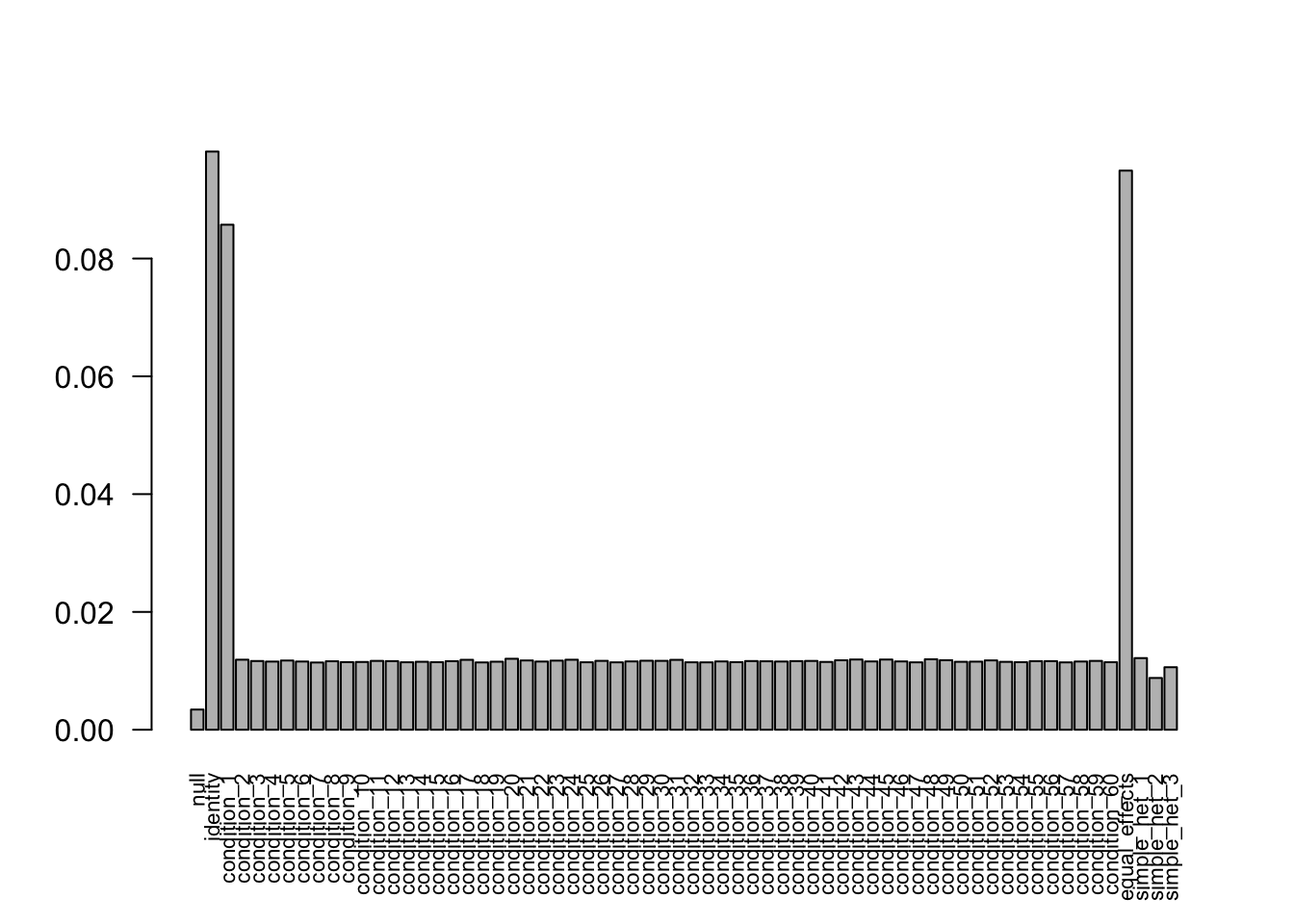
Without missing values
mash.data.missing.na.1 = mash_set_data(Bhat=data.miss.na$Bhat, Shat= data.miss.na$Shat, alpha = 1)
U.c = cov_canonical(mash.data.missing.na.1)
mash.model.missing.na.1 = mash(mash.data.missing.na.1, U.c, verbose=FALSE)Warning in REBayes::KWDual(A, rep(1, k), normalize(w), control = control): estimated mixing distribution has some negative values:
consider reducing rtolWarning in mixIP(matrix_lik = structure(c(0.240920767436896,
0.33756333012155, : Optimization step yields mixture weights that are
either too small, or negative; weights have been corrected and renormalized
after the optimization.FIXME: 'compute_posterior_matrices' in Rcpp does not transfer EZ to EEbarplot(get_estimated_pi(mash.model.missing.na.1), las=2, cex.names = 0.7)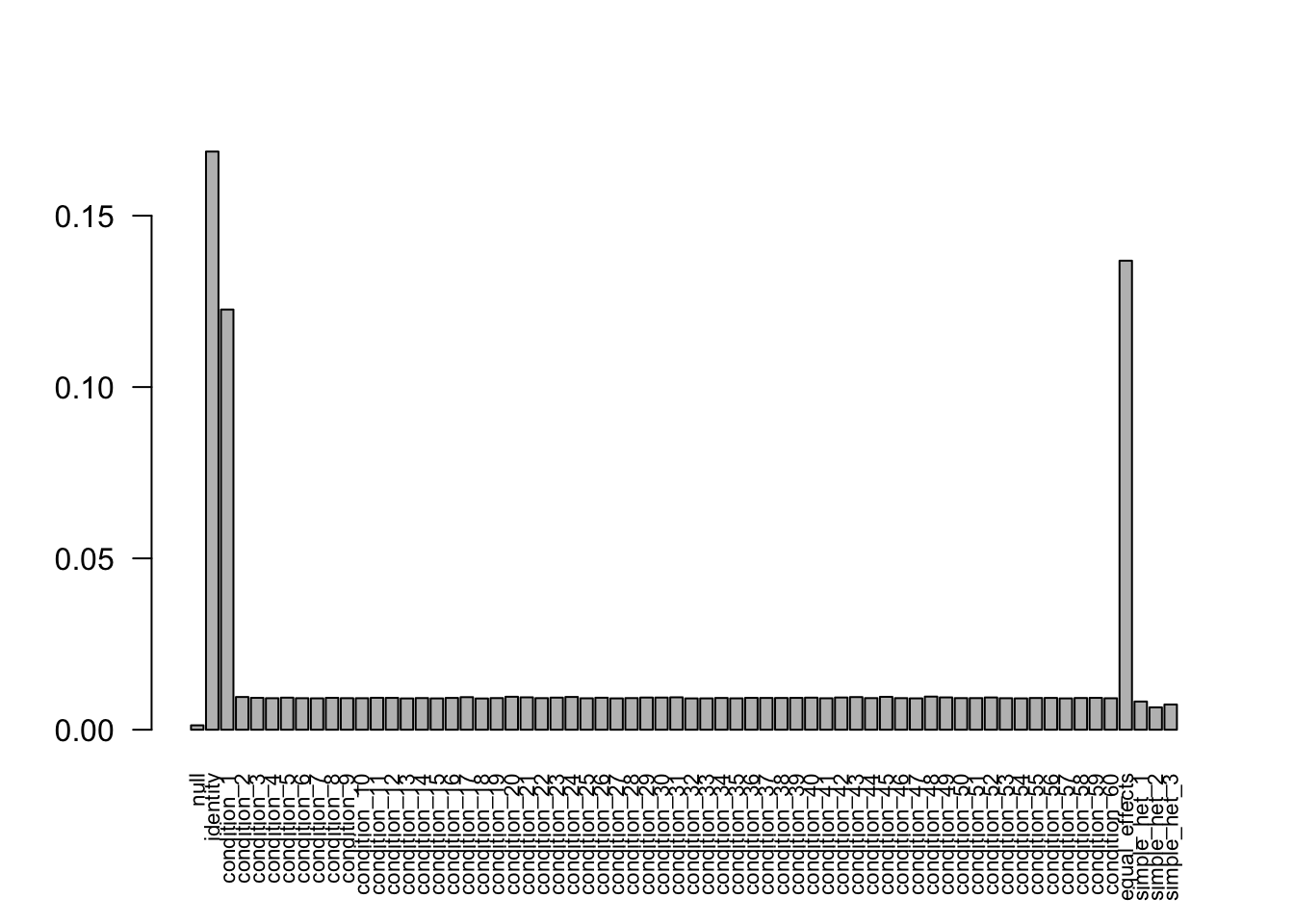 The estimated weights are different.
The estimated weights are different.
Subgroups
The weired covariance structure is caused by the high dimension of R.
If we separate conditions into several groups, the covariance structure will be correct.
We show this for the EZ model R=60.
FIXME: 'compute_posterior_matrices' in Rcpp does not transfer EZ to EE
FIXME: 'compute_posterior_matrices' in Rcpp does not transfer EZ to EE
FIXME: 'compute_posterior_matrices' in Rcpp does not transfer EZ to EE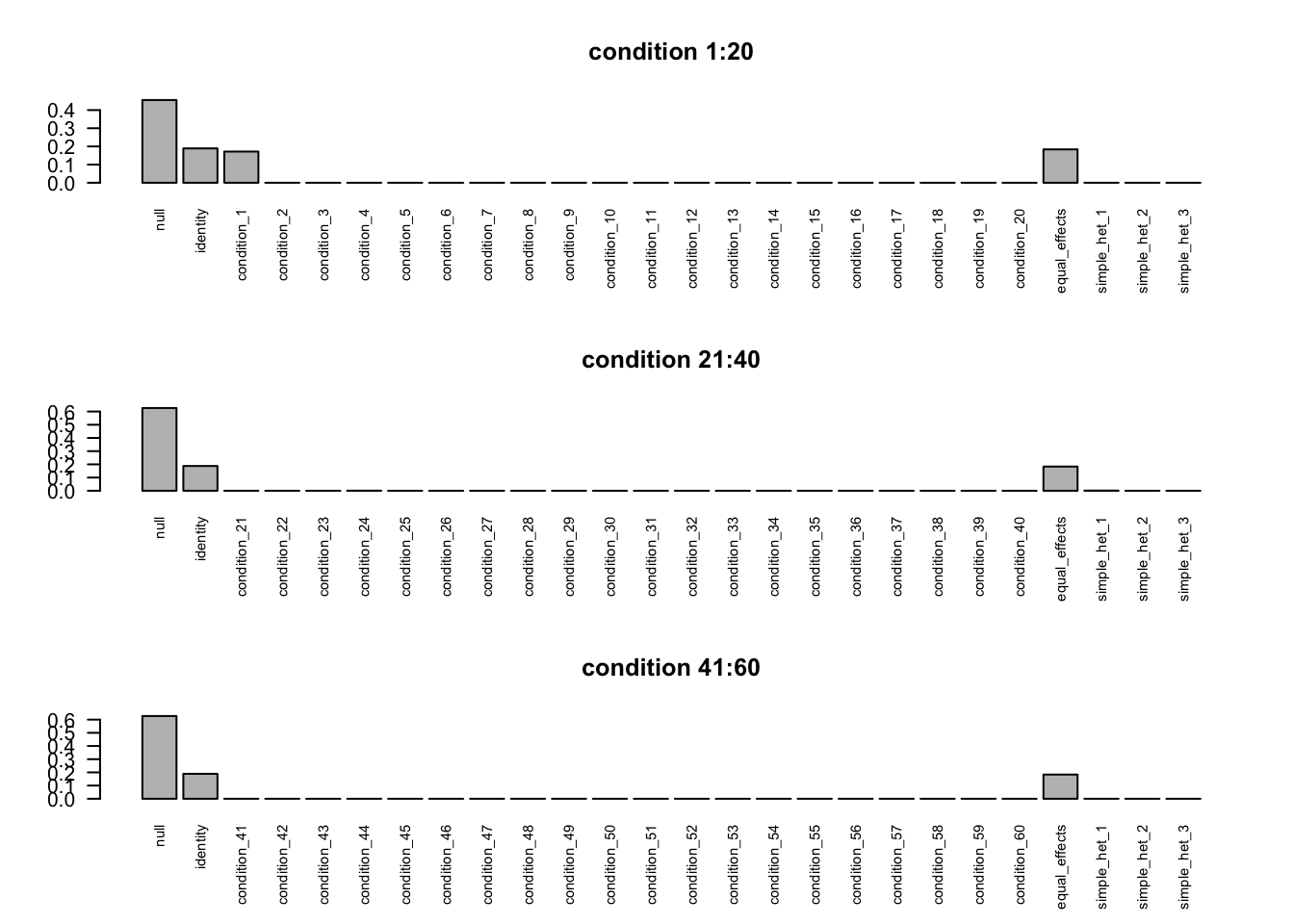
Session information
sessionInfo()R version 3.4.3 (2017-11-30)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS High Sierra 10.13.3
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] mashr_0.2-6 ashr_2.2-7
loaded via a namespace (and not attached):
[1] Rcpp_0.12.15 knitr_1.20 magrittr_1.5
[4] REBayes_1.2 MASS_7.3-47 doParallel_1.0.11
[7] pscl_1.5.2 SQUAREM_2017.10-1 lattice_0.20-35
[10] foreach_1.4.4 plyr_1.8.4 stringr_1.3.0
[13] tools_3.4.3 parallel_3.4.3 grid_3.4.3
[16] rmeta_2.16 git2r_0.20.0 htmltools_0.3.6
[19] iterators_1.0.9 assertthat_0.2.0 yaml_2.1.17
[22] rprojroot_1.2 digest_0.6.13 Matrix_1.2-12
[25] codetools_0.2-15 evaluate_0.10.1 rmarkdown_1.8
[28] stringi_1.1.6 compiler_3.4.3 Rmosek_8.0.69
[31] backports_1.1.2 mvtnorm_1.0-7 truncnorm_1.0-8 This R Markdown site was created with workflowr