Check read counts in binned genome
Briana Mittleman
2017-11-30
Last updated: 2017-11-30
Code version: ab9d530
Bash script
Split genome into 200bp windows and run the coverage command:
/project2/gilad/briana/Net-seq/genome_bed/hg19_200bp_window.bed
/project2/gilad/briana/Net-seq/ref_genes/windows_200
make window_200_cov.sh
#!/bin/bash
#SBATCH --job-name=window_200_cov
#SBATCH --time=8:00:00
#SBATCH --partition=broadwl
#SBATCH --mem=50G
#SBATCH --tasks-per-node=4
#SBATCH --mail-type=END
bedtools coverage -counts -a /project2/gilad/briana/Net-seq/genome_bed/hg19_200bp_window.bed -b /project2/gilad/briana/Net-seq/Net-seq1/data/bed_sort/net1_18486_dep_chr_sort.bed > /project2/gilad/briana/Net-seq/ref_genes/windows_200/window_200_cov_18486.txt
bedtools coverage -counts -a /project2/gilad/briana/Net-seq/genome_bed/hg19_200bp_window.bed -b /project2/gilad/briana/Net-seq/Net-seq1/data/bed_sort/net1_18508_dep_chr_sort.bed > /project2/gilad/briana/Net-seq/ref_genes/windows_200/window_200_cov_18508_dep.txt
bedtools coverage -counts -a /project2/gilad/briana/Net-seq/genome_bed/hg19_200bp_window.bed -b /project2/gilad/briana/Net-seq/Net-seq1/data/bed_sort/net1_18508_nondep_chr_sort.bed > /project2/gilad/briana/Net-seq/ref_genes/windows_200/window_200_cov_18508_nondep.txt
bedtools coverage -counts -a /project2/gilad/briana/Net-seq/genome_bed/hg19_200bp_window.bed -b /project2/gilad/briana/Net-seq/Net-seq1/data/bed_sort/net1_19238_dep_chr_sort.bed > /project2/gilad/briana/Net-seq/ref_genes/windows_200/window_200_cov_19238.txt
bedtools coverage -counts -a /project2/gilad/briana/Net-seq/genome_bed/hg19_200bp_window.bed -b /project2/gilad/briana/Net-seq/data/bed_sort/mayer_SRR1575922_chr_sort.bed > /project2/gilad/briana/Net-seq/ref_genes/windows_200/window_200_cov_mayer.txt
#bedtools coverage -counts -a /project2/gilad/briana/Net-seq/genome_bed/hg19_200bp_window.bed -b /project2/gilad/briana/Net-seq/Net-seq1/data/bed/merged_Net1.chr.bed > /project2/gilad/briana/Net-seq/ref_genes/windows_200/window_200_cov_merged.txt
#step memory exceeded!Import data
window_200_18486=read.csv("../data/windows_200/window_200_cov_18486.txt", header=FALSE, sep="\t")
window_200_18508_dep=read.csv("../data/windows_200/window_200_cov_18508_dep.txt", header=FALSE, sep="\t")
window_200_18508_nondep=read.csv("../data/windows_200/window_200_cov_18508_nondep.txt", header=FALSE, sep="\t")
window_200_19238=read.csv("../data/windows_200/window_200_cov_19238.txt", header=FALSE, sep="\t")
window_200_mayer= read.csv("../data/windows_200/window_200_cov_mayer.txt", header=FALSE, sep="\t")Add col labels to each file:
colnames(window_200_18486) = c("chr", "start", "end", "count")
colnames(window_200_18508_dep) = c("chr", "start", "end", "count")
colnames(window_200_18508_nondep) = c("chr", "start", "end", "count")
colnames(window_200_19238) = c("chr", "start", "end", "count")
colnames(window_200_mayer) = c("chr", "start", "end", "count")Plot data
Data I want to look at:
- summary per library
summary(window_200_18486$count) Min. 1st Qu. Median Mean 3rd Qu. Max.
0 0 0 1 0 4076520 summary(window_200_18508_dep$count) Min. 1st Qu. Median Mean 3rd Qu. Max.
0 0 0 21 0 27069584 summary(window_200_18508_nondep$count) Min. 1st Qu. Median Mean 3rd Qu. Max.
0 0 0 23 0 30571781 summary(window_200_19238$count) Min. 1st Qu. Median Mean 3rd Qu. Max.
0 0 0 4 0 13033253 summary(window_200_mayer$count) Min. 1st Qu. Median Mean 3rd Qu. Max.
0.0 0.0 0.0 113.3 0.0 701170.0 Use a plot to see the distribution:
- summaries not including zero
Make dataframe excluding the zeros:
window_200_18486_non0= window_200_18486[window_200_18486$count!=0,]
window_200_18508_dep_non0= window_200_18508_dep[window_200_18508_dep$count!=0,]
window_200_18508_nondep_non0= window_200_18508_nondep[window_200_18508_nondep$count!=0,]
window_200_19238_non0= window_200_19238[window_200_19238$count!=0,]
window_200_mayer_non0= window_200_mayer[window_200_mayer$count!=0,]summarise
summary(window_200_18486_non0$count) Min. 1st Qu. Median Mean 3rd Qu. Max.
1 1 1 45 3 4076520 summary(window_200_18508_dep_non0$count) Min. 1st Qu. Median Mean 3rd Qu. Max.
1 1 1 624 3 27069584 summary(window_200_18508_nondep_non0$count) Min. 1st Qu. Median Mean 3rd Qu. Max.
1 1 1 633 3 30571781 summary(window_200_19238_non0$count) Min. 1st Qu. Median Mean 3rd Qu. Max.
1 1 1 149 3 13033253 summary(window_200_mayer_non0$count) Min. 1st Qu. Median Mean 3rd Qu. Max.
1 1 1 2200 2 701170 plot(sort(log(window_200_19238_non0$count), decreasing=TRUE))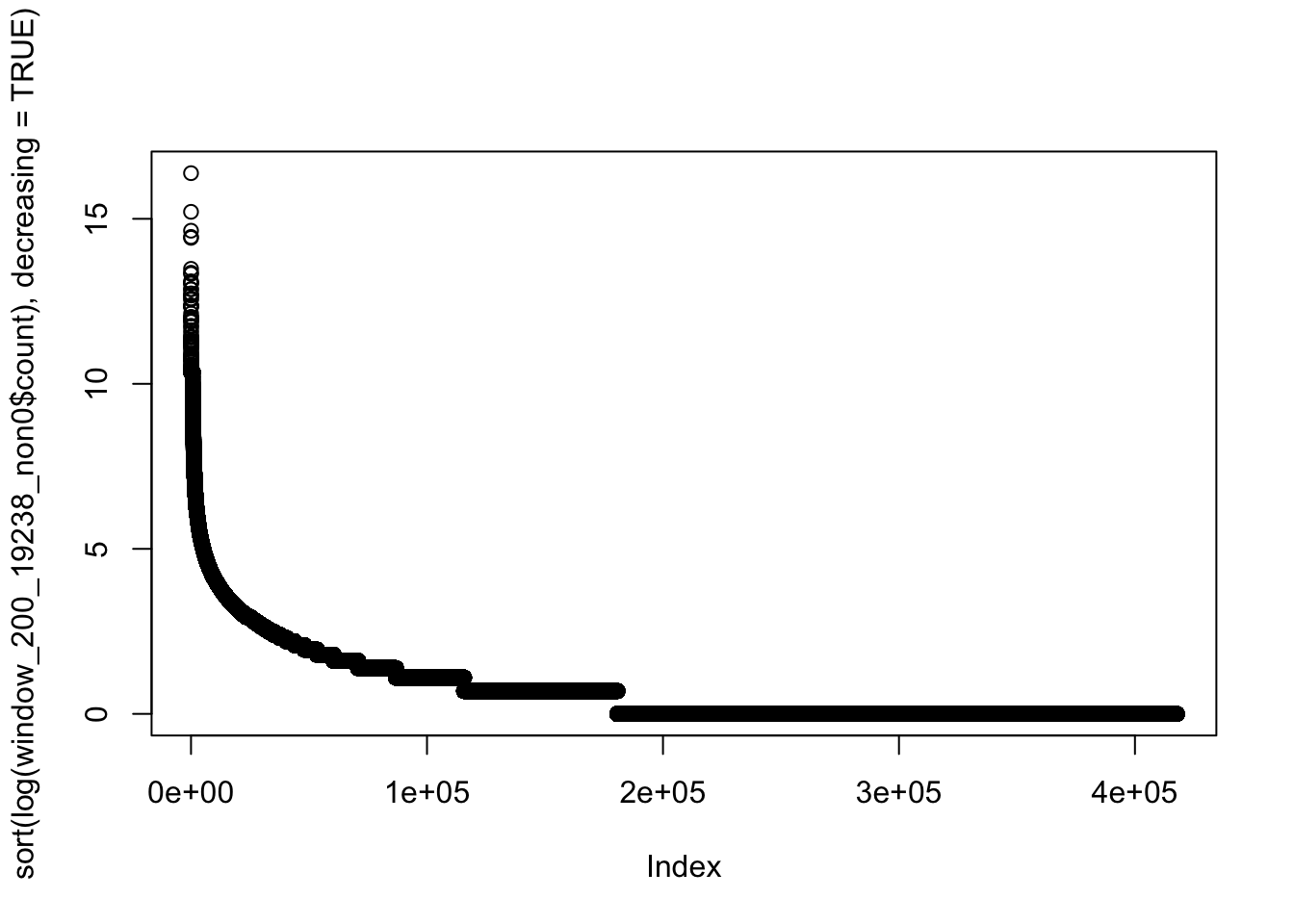
- number of entries that are non zero
x= nrow(window_200_18486)
barplot(c(nrow(window_200_18486_non0)/x,nrow(window_200_18508_dep_non0)/x,nrow(window_200_18508_nondep_non0)/x, nrow(window_200_19238_non0)/x), main="Proportion of detected bins", names=c("18486", "18508 \n dep", "18508 \n nondep", "19238"))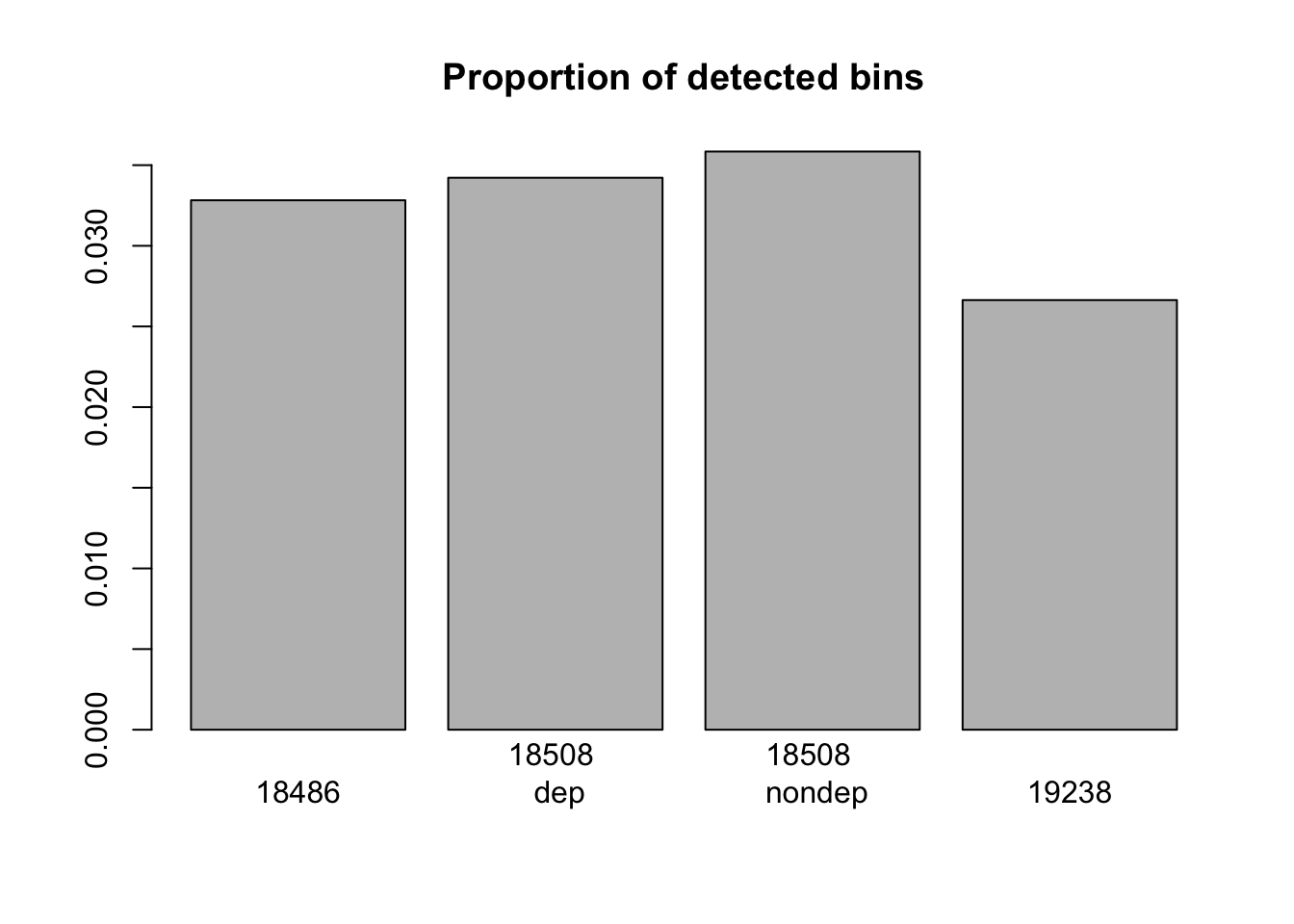
nrow(window_200_mayer_non0)/x[1] 0.05149546Session information
sessionInfo()R version 3.4.2 (2017-09-28)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS Sierra 10.12.6
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
loaded via a namespace (and not attached):
[1] compiler_3.4.2 backports_1.1.1 magrittr_1.5 rprojroot_1.2
[5] tools_3.4.2 htmltools_0.3.6 yaml_2.1.14 Rcpp_0.12.13
[9] stringi_1.1.5 rmarkdown_1.6 knitr_1.17 git2r_0.19.0
[13] stringr_1.2.0 digest_0.6.12 evaluate_0.10.1This R Markdown site was created with workflowr