Implementation by Rmosek: Correlated Null
Lei Sun
2017-05-02
Last updated: 2017-11-07
Code version: 2c05d59
Introduction
Being in the development stage, cvxr is not very efficient at the convex optimization in fitting Gaussian derivatives. We are moving to Rmosek.
source("../code/gdash.R")Warning: replacing previous import 'Matrix::crossprod' by 'gmp::crossprod'
when loading 'cvxr'Warning: replacing previous import 'Matrix::tcrossprod' by
'gmp::tcrossprod' when loading 'cvxr'z.mat = read.table("../output/z_null_liver_777.txt")
fd.bh = scan("../output/fd.bh.0.05_null_liver_777.txt")
pihat0 = scan("../output/pihat0_z_null_liver_777.txt")Bad
These data sets are unfriendly for both BH and ASH.
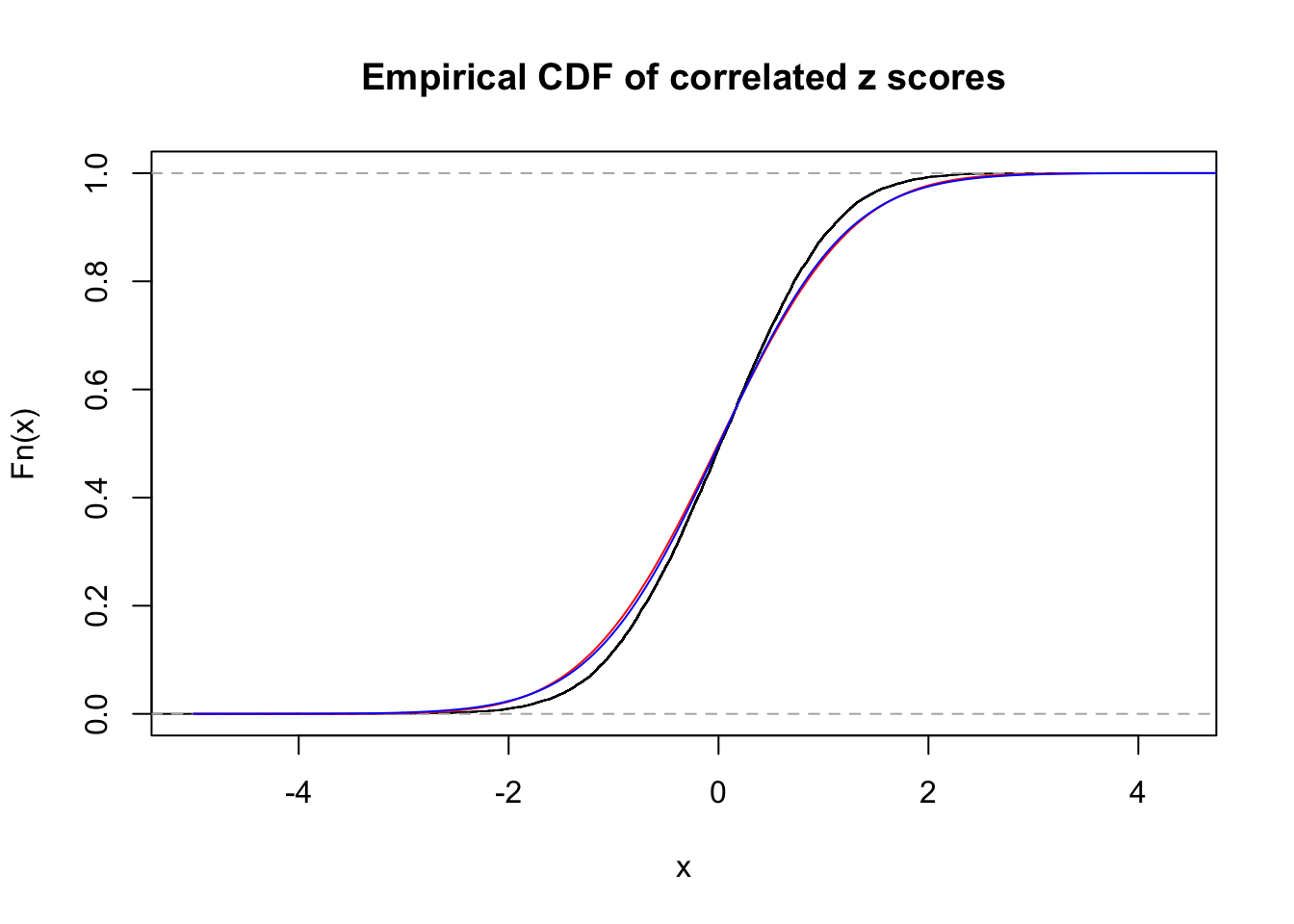
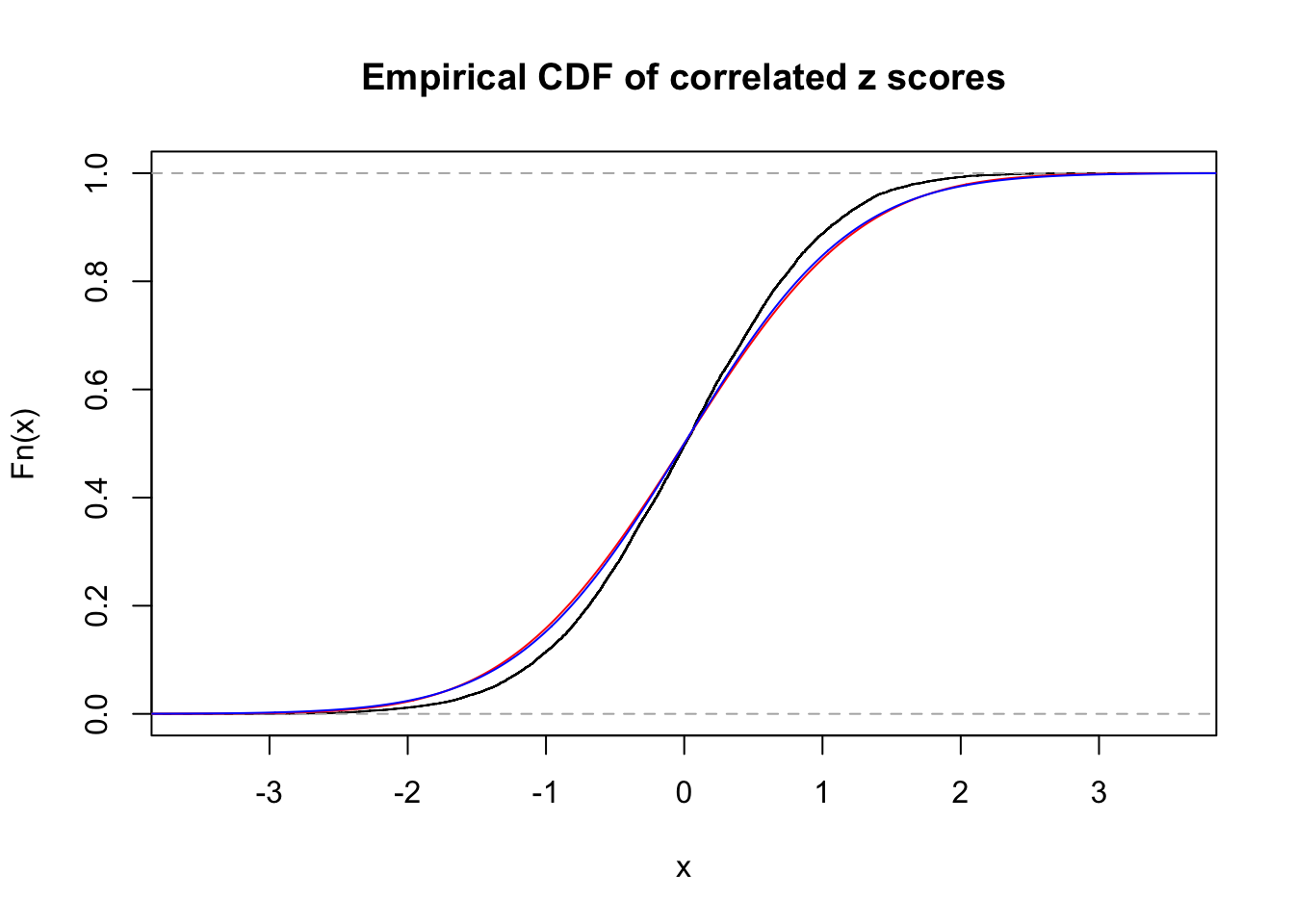
Data Set 355 :
Number of BH false discoveries: 639 ;
ASH's pihat0 = 0.04750946 ;
Normalized coefficients of Gaussian derivatives:
1 : 0.0225585778660563 ; 2 : 1.30307541409737 ; 3 : 0.0529119988564207 ; 4 : 0.86434625413445 ; 5 : 0 ; 6 : 0.115991285668782 ; 7 : 0 ; 8 : 0 ; 9 : 0 ; 10 : 0.012768069558034 ;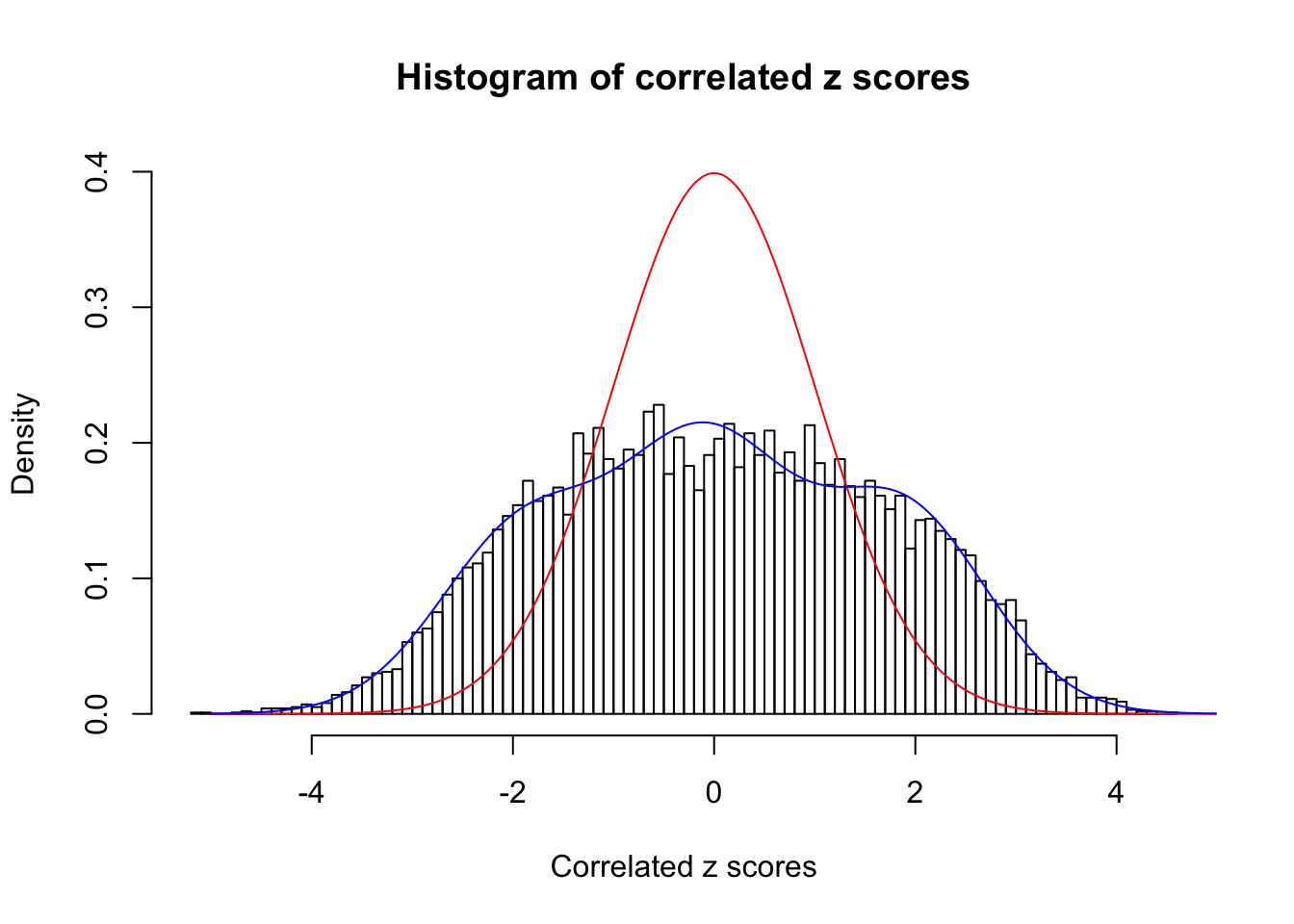
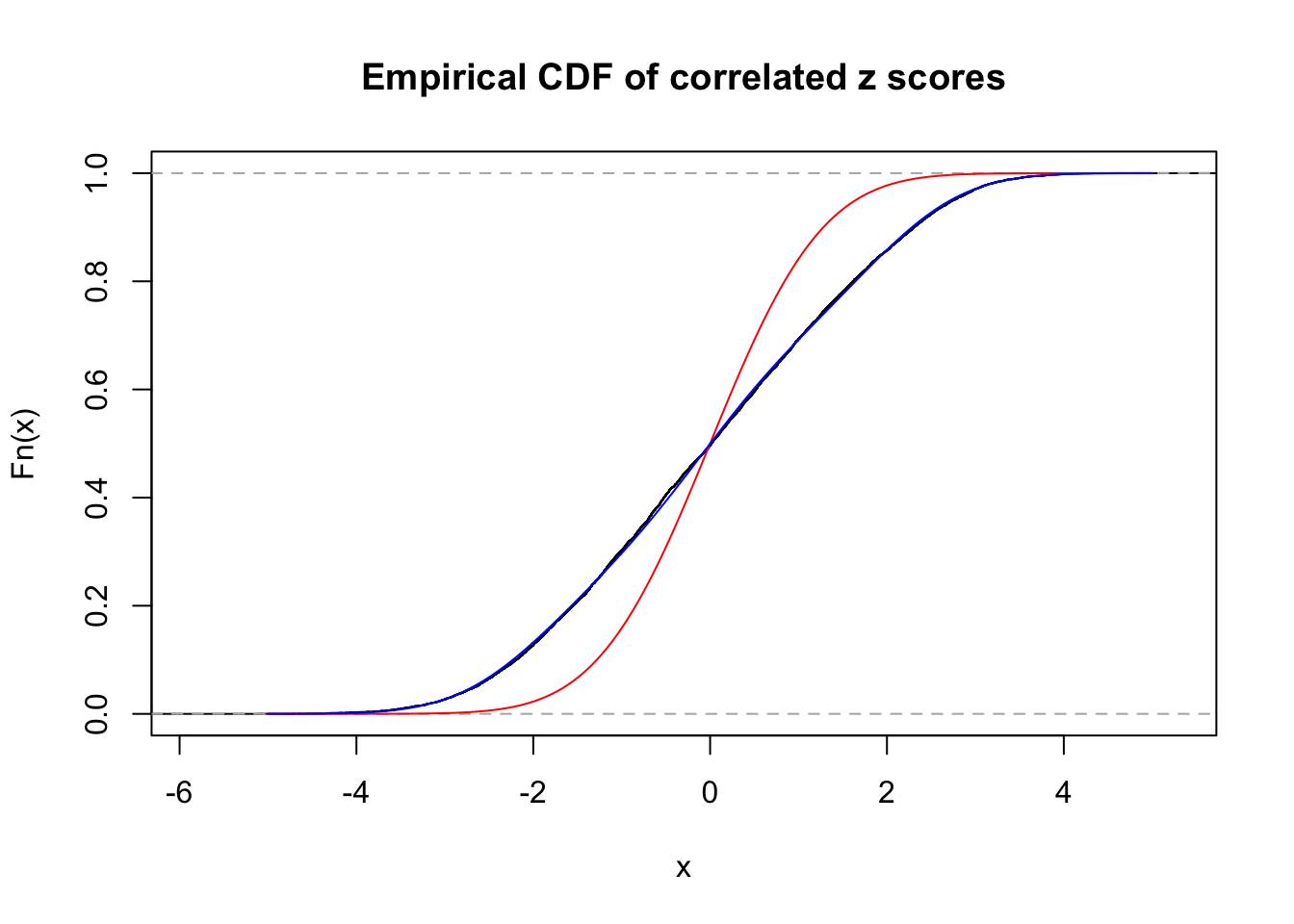
Data Set 23 :
Number of BH false discoveries: 408 ;
ASH's pihat0 = 0.03139009 ;
Normalized coefficients of Gaussian derivatives:
1 : 0.016039132567407 ; 2 : 1.34993003351774 ; 3 : 0.081263929095916 ; 4 : 0.714486051374311 ; 5 : 0.139320458700019 ; 6 : 0 ; 7 : 0.100823265137825 ; 8 : 0 ; 9 : 0.0608462191710317 ; 10 : 0.000964802431909129 ;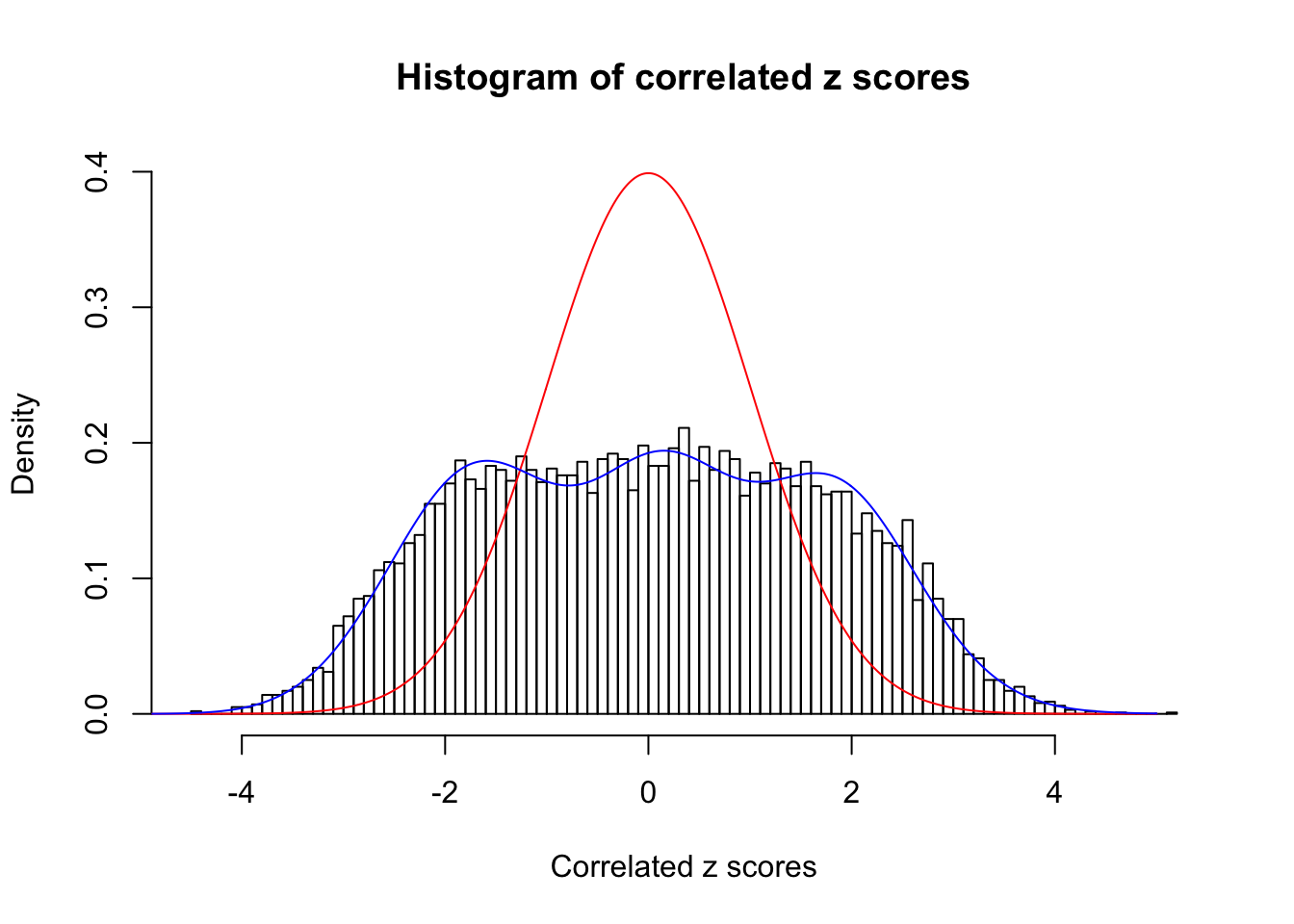
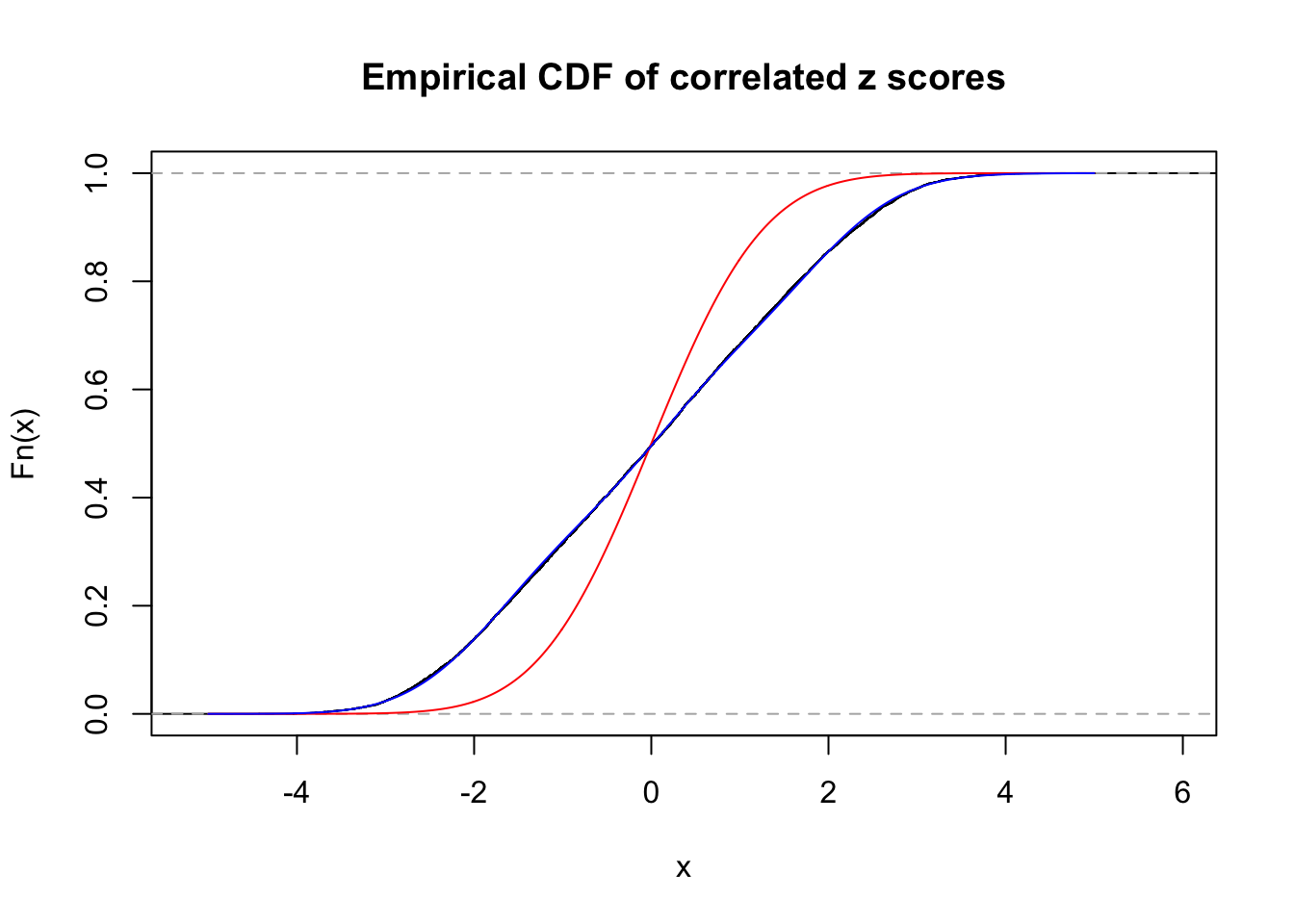
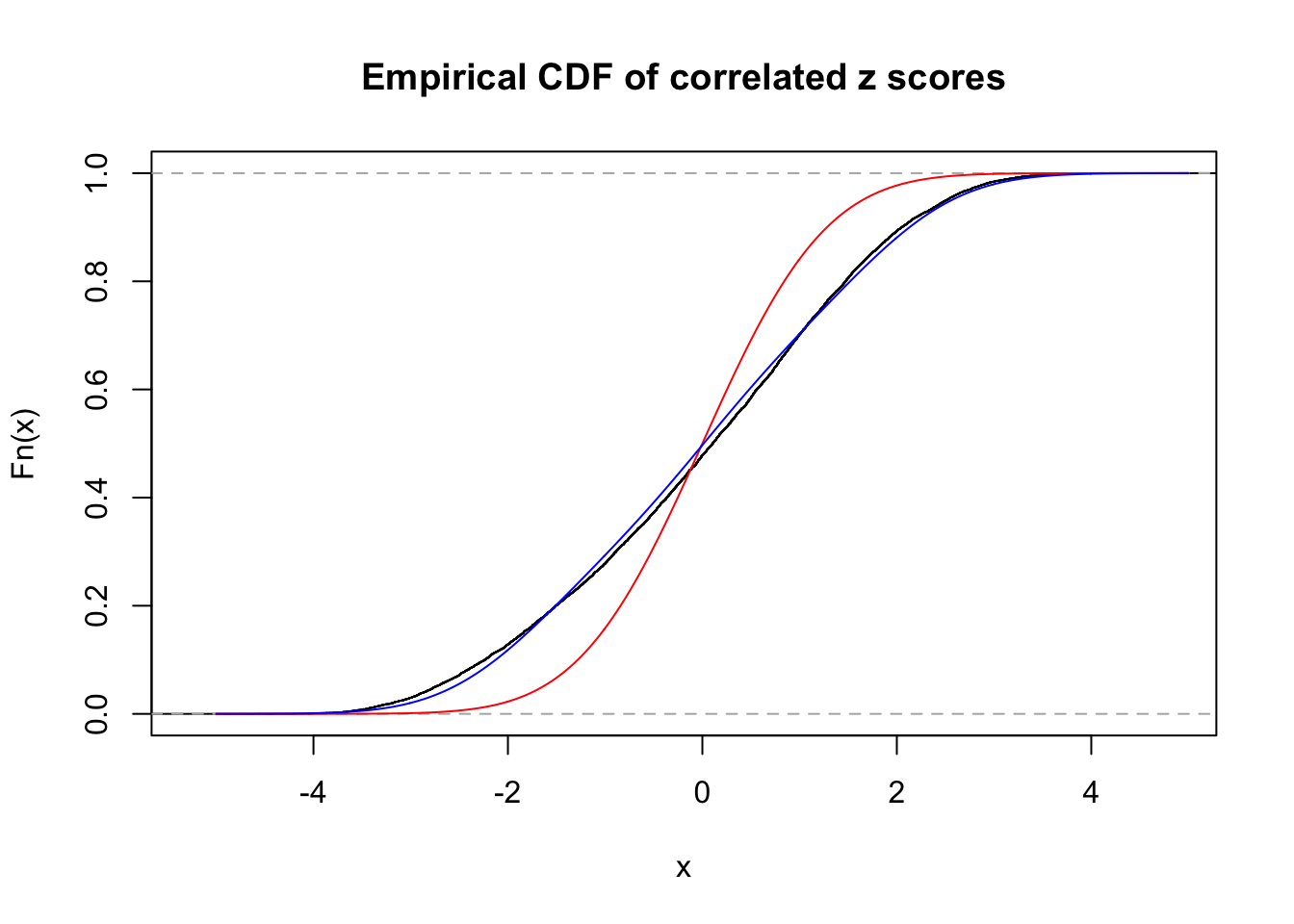
Data Set 122 :
Number of BH false discoveries: 331 ;
ASH's pihat0 = 0.04301549 ;
Normalized coefficients of Gaussian derivatives:
1 : 0.00711459304239053 ; 2 : 1.1037821799492 ; 3 : 0 ; 4 : 0.528402132228434 ; 5 : 0 ; 6 : 0 ; 7 : 0 ; 8 : 0 ; 9 : 0.000977244957534518 ; 10 : 0 ;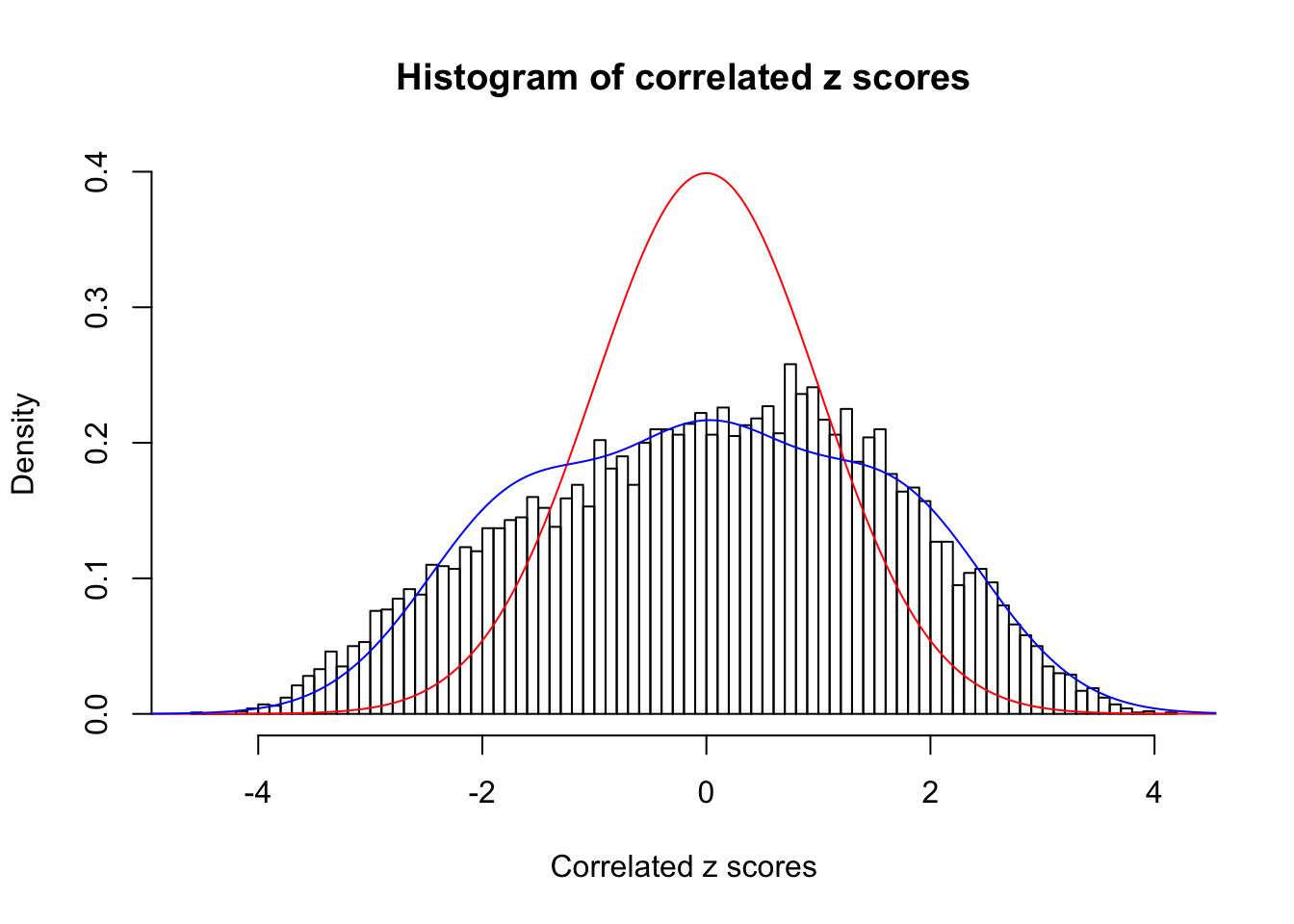
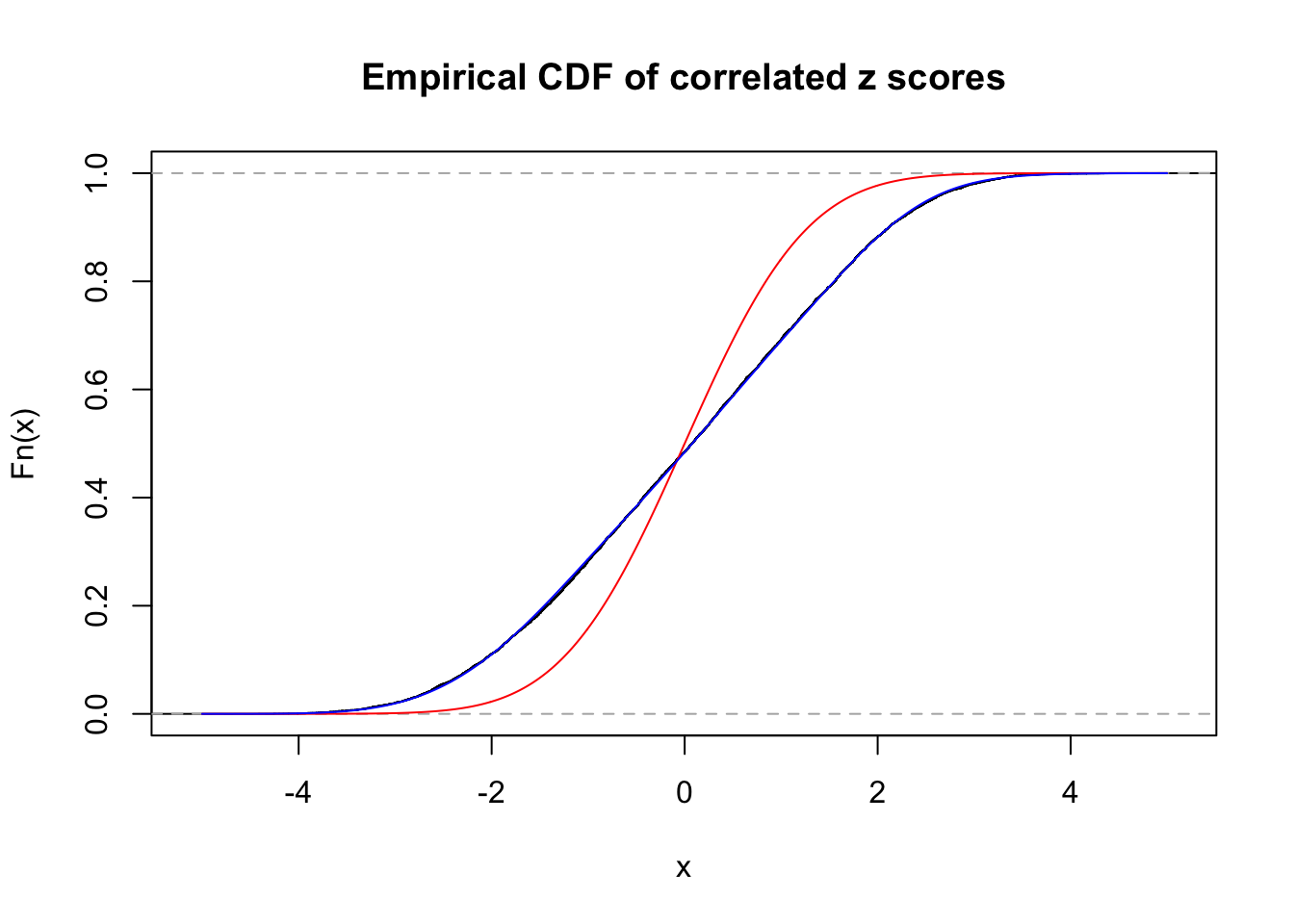
Data Set 749 :
Number of BH false discoveries: 114 ;
ASH's pihat0 = 0.02583276 ;
Normalized coefficients of Gaussian derivatives:
1 : 0.0458626422285871 ; 2 : 1.0692347200711 ; 3 : 0.00127673630293041 ; 4 : 0.437875595583542 ; 5 : 0 ; 6 : 0 ; 7 : 0.110561067339144 ; 8 : 0 ; 9 : 0.104817311821469 ; 10 : 0 ;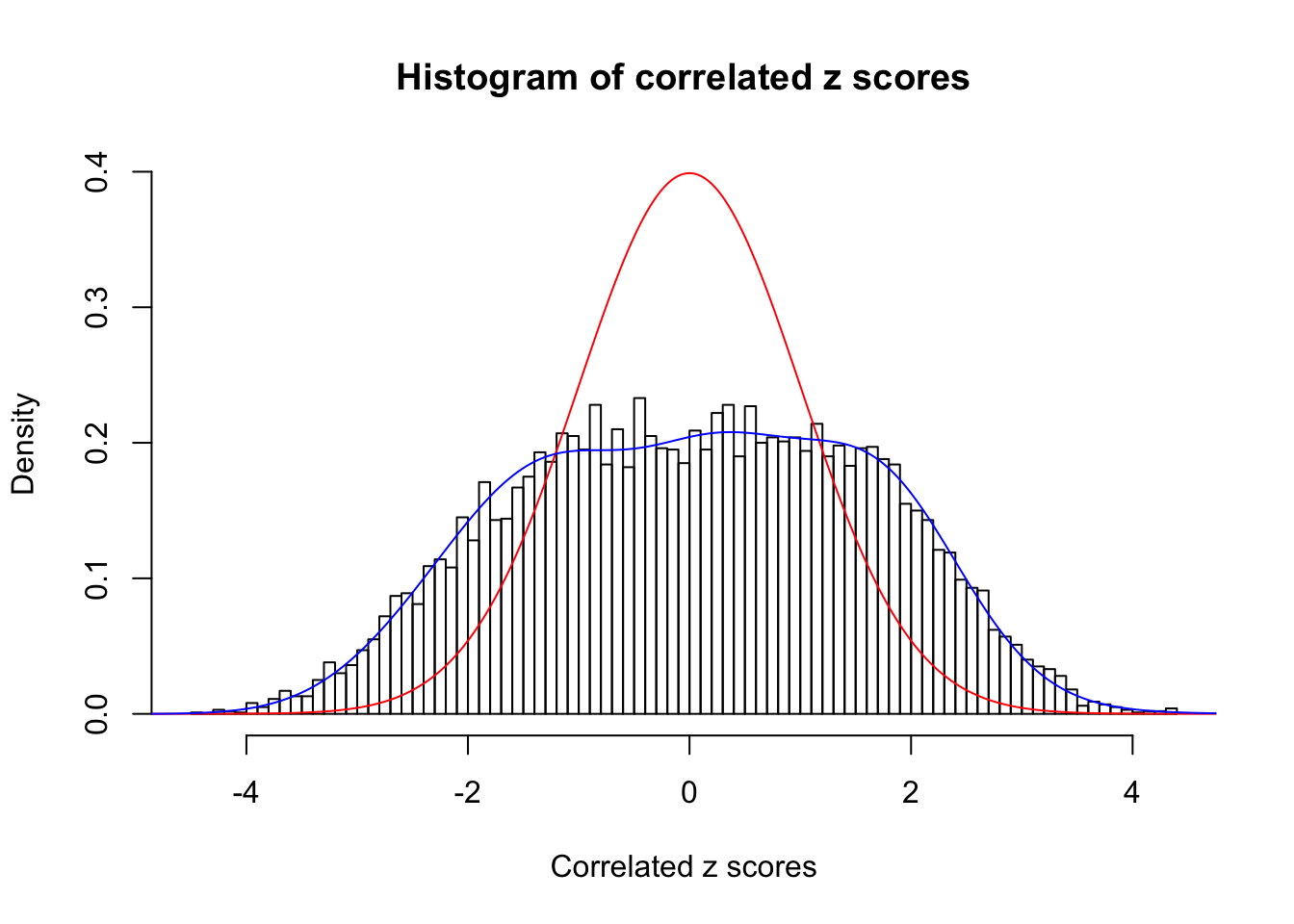
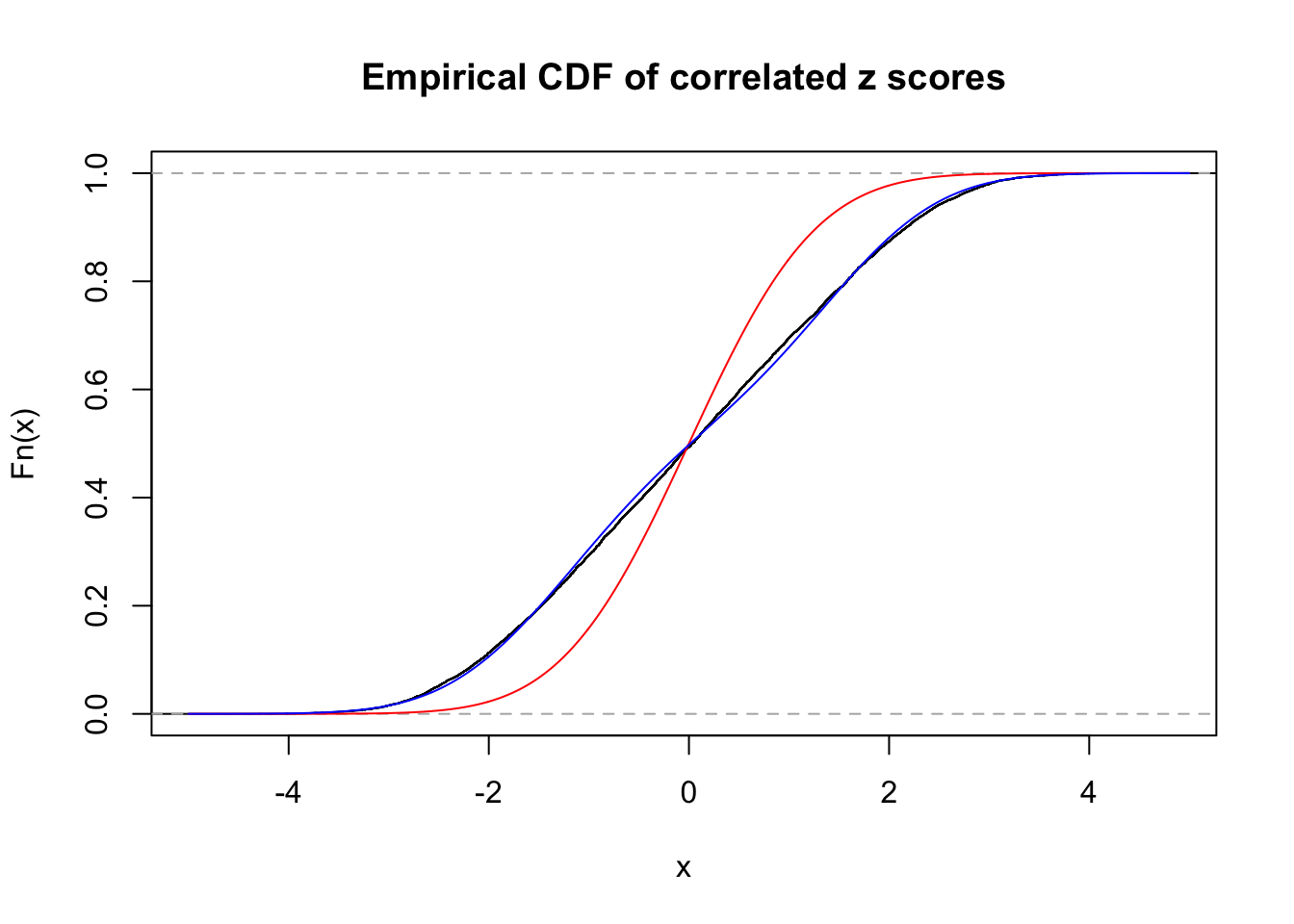
Data Set 724 :
Number of BH false discoveries: 79 ;
ASH's pihat0 = 0.01606004 ;
Normalized coefficients of Gaussian derivatives:
1 : 0.0352454327180146 ; 2 : 1.085621208483 ; 3 : 0.072031313135509 ; 4 : 0.313477034131008 ; 5 : 0 ; 6 : 0 ; 7 : 0 ; 8 : 0 ; 9 : 0 ; 10 : 0 ;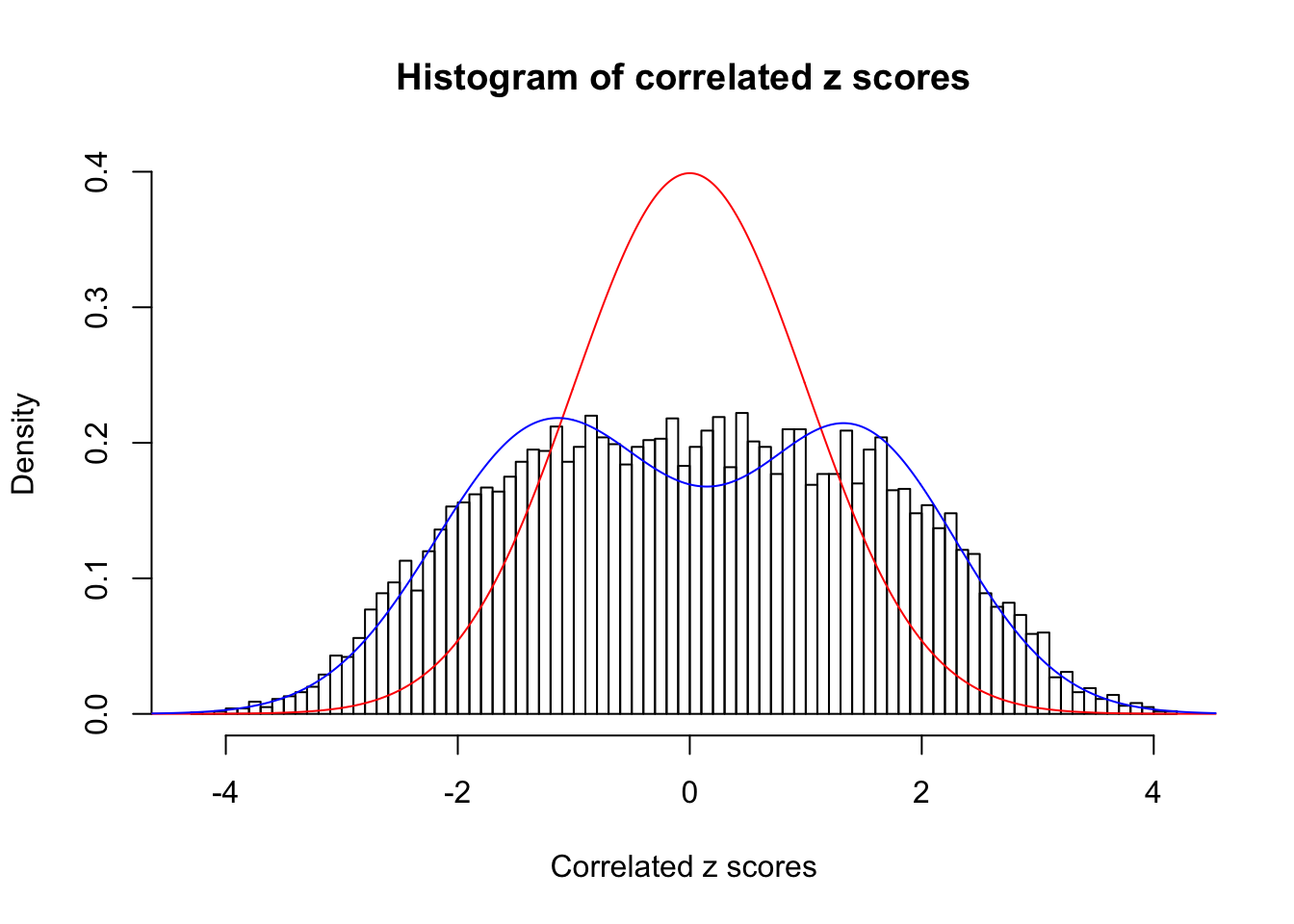
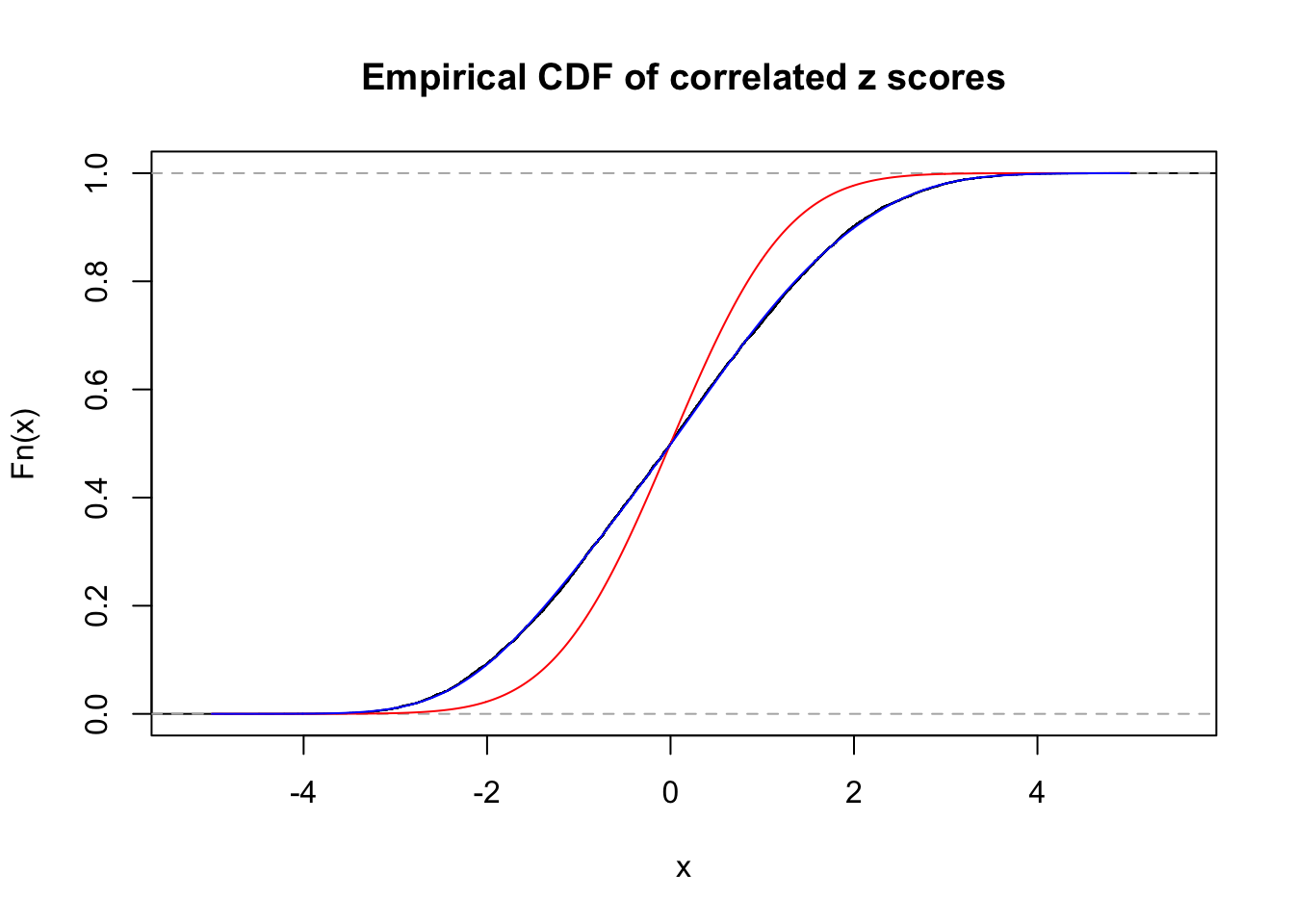
Data Set 56 :
Number of BH false discoveries: 35 ;
ASH's pihat0 = 0.04829662 ;
Normalized coefficients of Gaussian derivatives:
1 : 0.015603437988185 ; 2 : 0.863695602978145 ; 3 : 0.135238823162622 ; 4 : 0.336622180491709 ; 5 : 0.18630322261983 ; 6 : 0.00268136598387189 ; 7 : 0.0339747362866559 ; 8 : 0.0181307043838827 ; 9 : 0 ; 10 : 0.0344310653486728 ;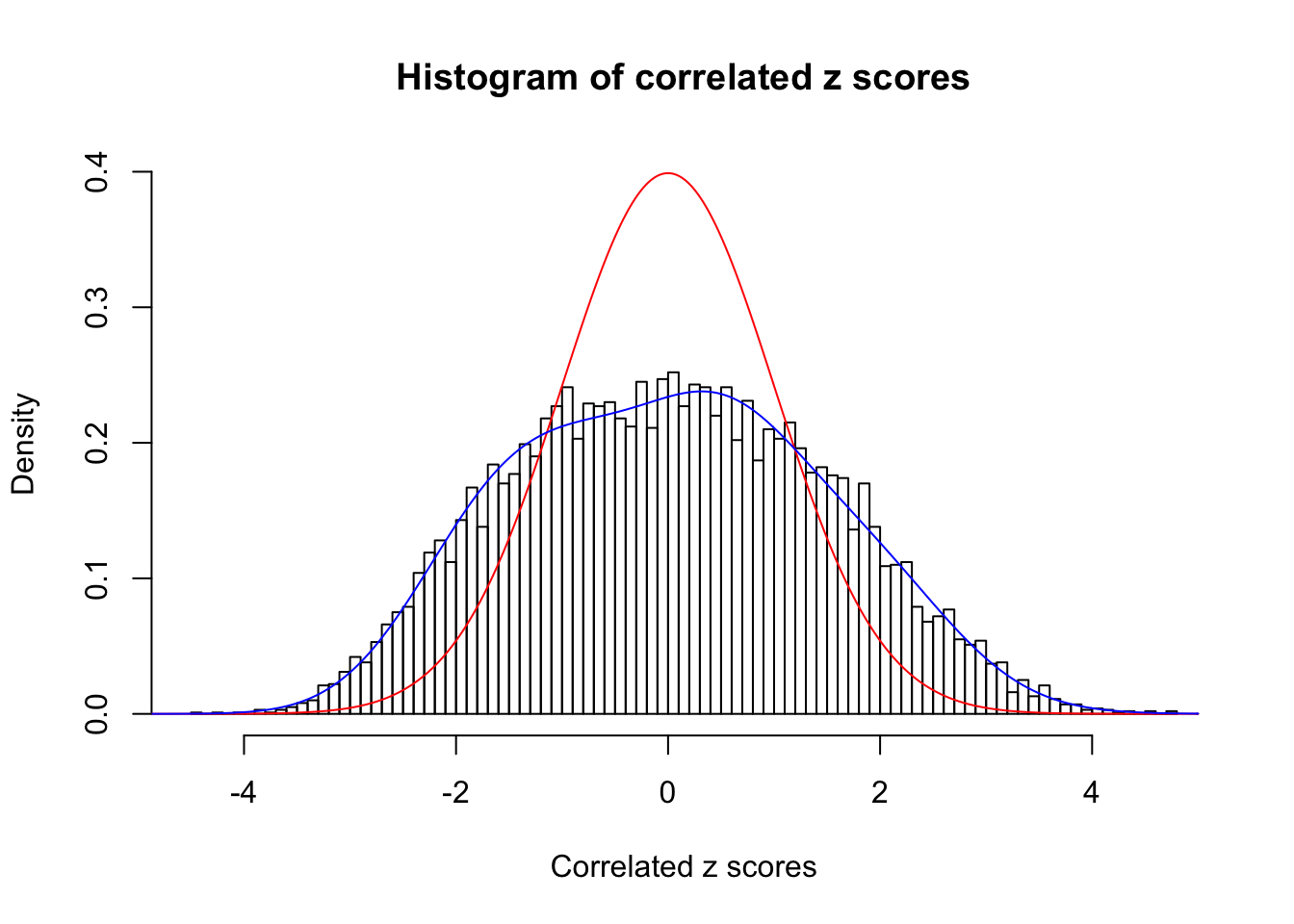
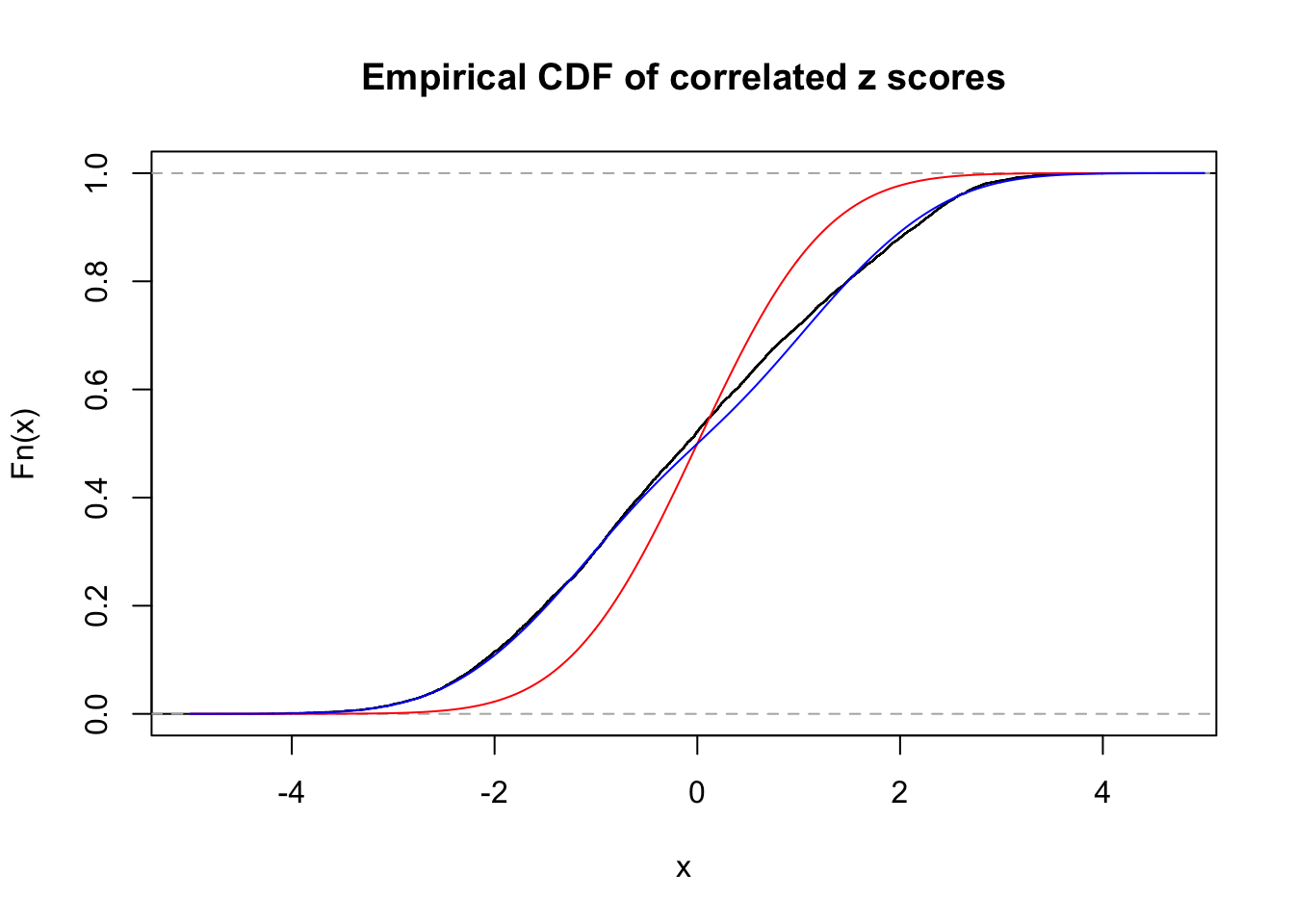
Data Set 840 :
Number of BH false discoveries: 28 ;
ASH's pihat0 = 0.02316832 ;
Normalized coefficients of Gaussian derivatives:
1 : 0 ; 2 : 1.03471486197017 ; 3 : 0 ; 4 : 0.329235042804543 ; 5 : 0 ; 6 : 0 ; 7 : 0 ; 8 : 0 ; 9 : 0.00321461375022215 ; 10 : 0.0593306511691146 ;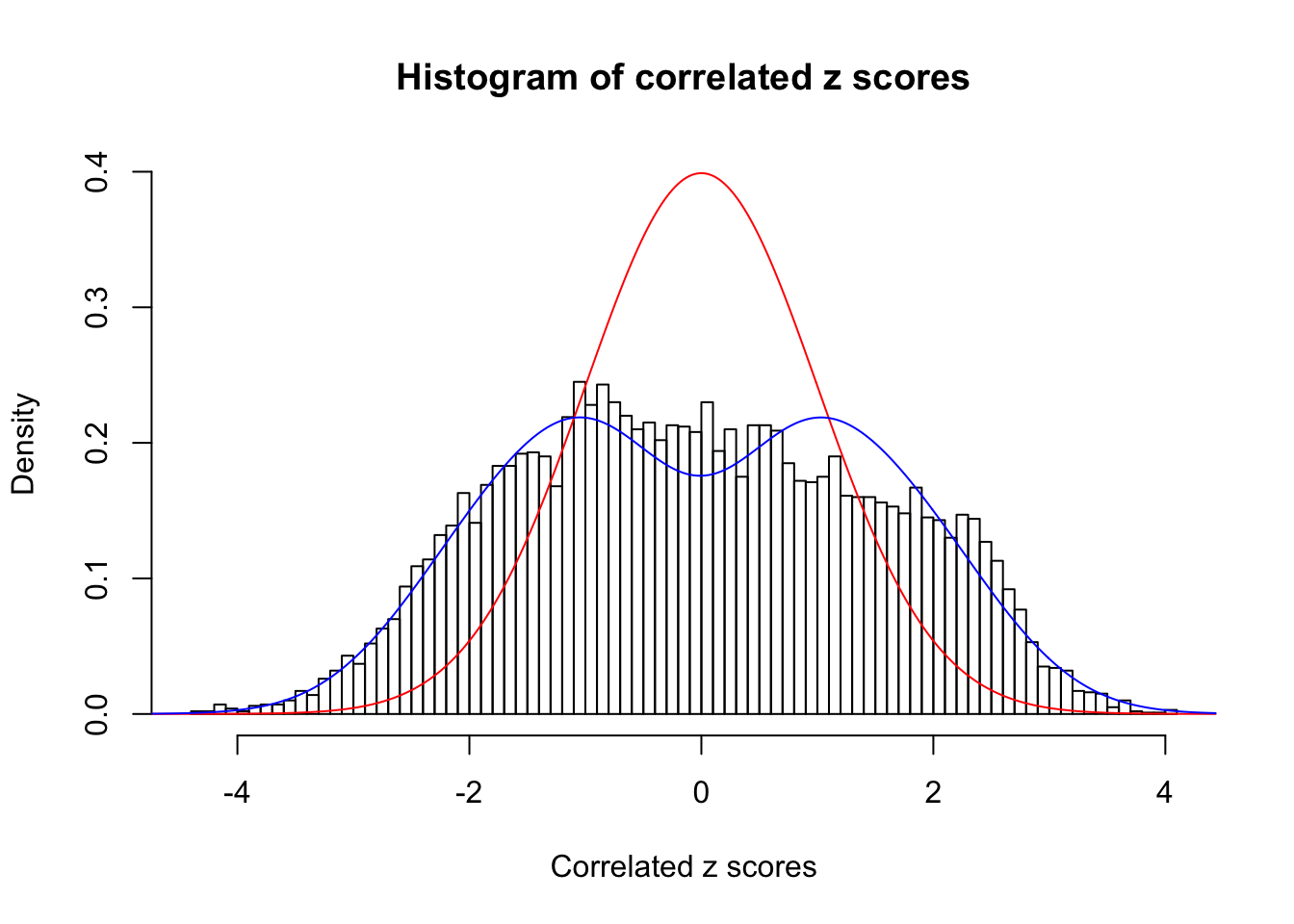
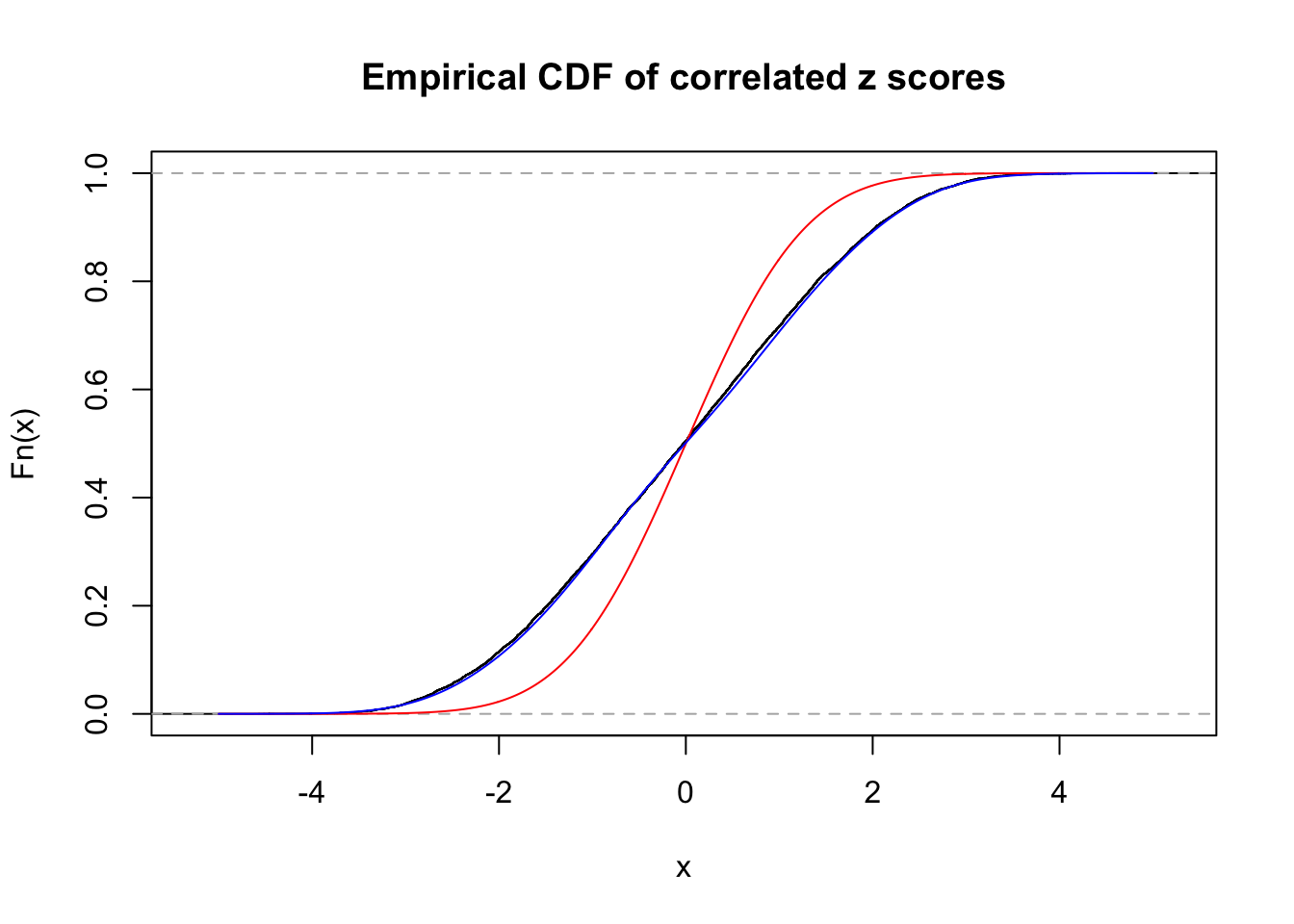
Data Set 858 :
Number of BH false discoveries: 16 ;
ASH's pihat0 = 0.05110069 ;
Normalized coefficients of Gaussian derivatives:
1 : 0 ; 2 : 1.00711281742885 ; 3 : 0 ; 4 : 0.390477482047559 ; 5 : 0 ; 6 : 0 ; 7 : 0.0551377637495718 ; 8 : 0 ; 9 : 0.0479292544081263 ; 10 : 0.0837572023674743 ;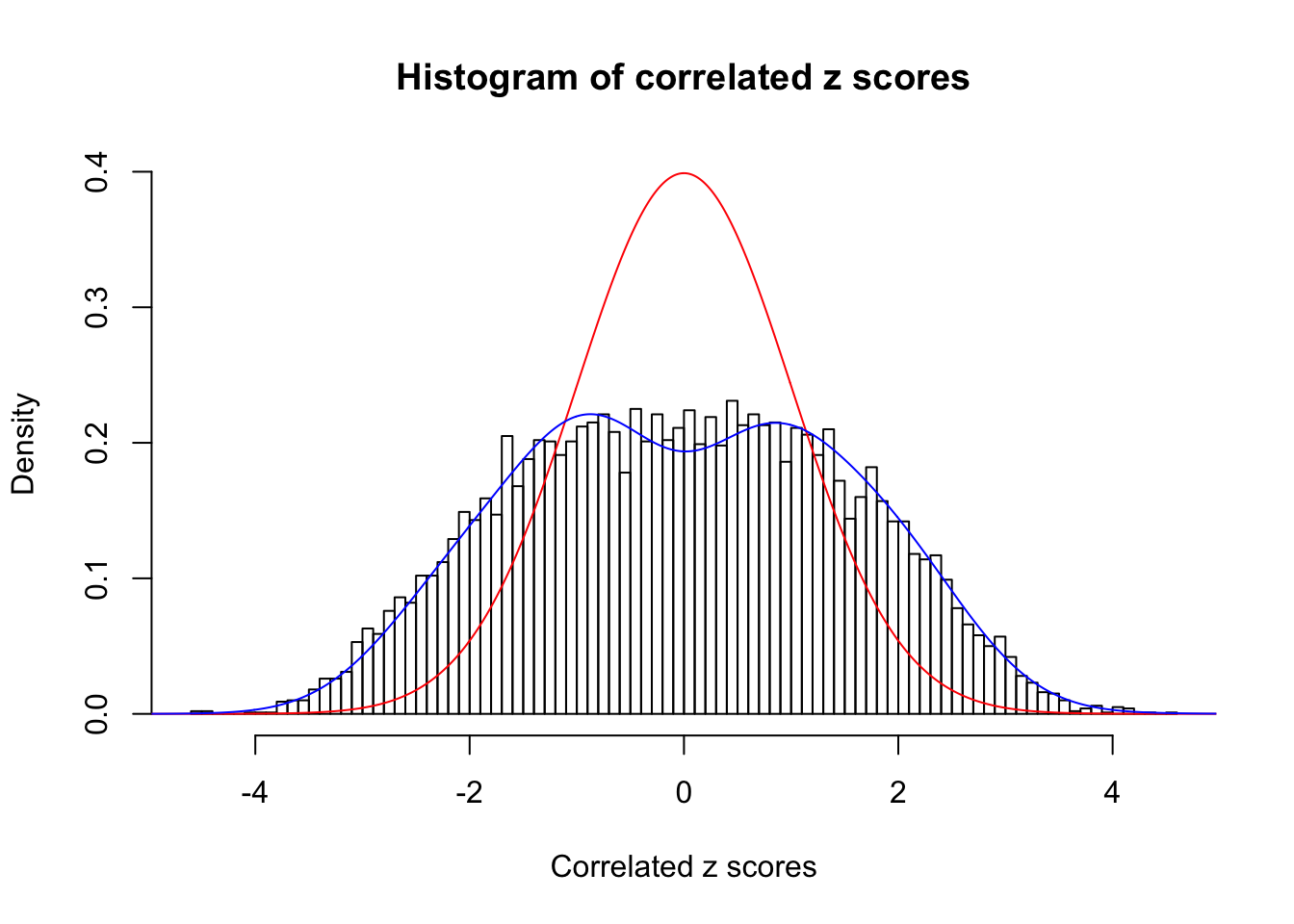
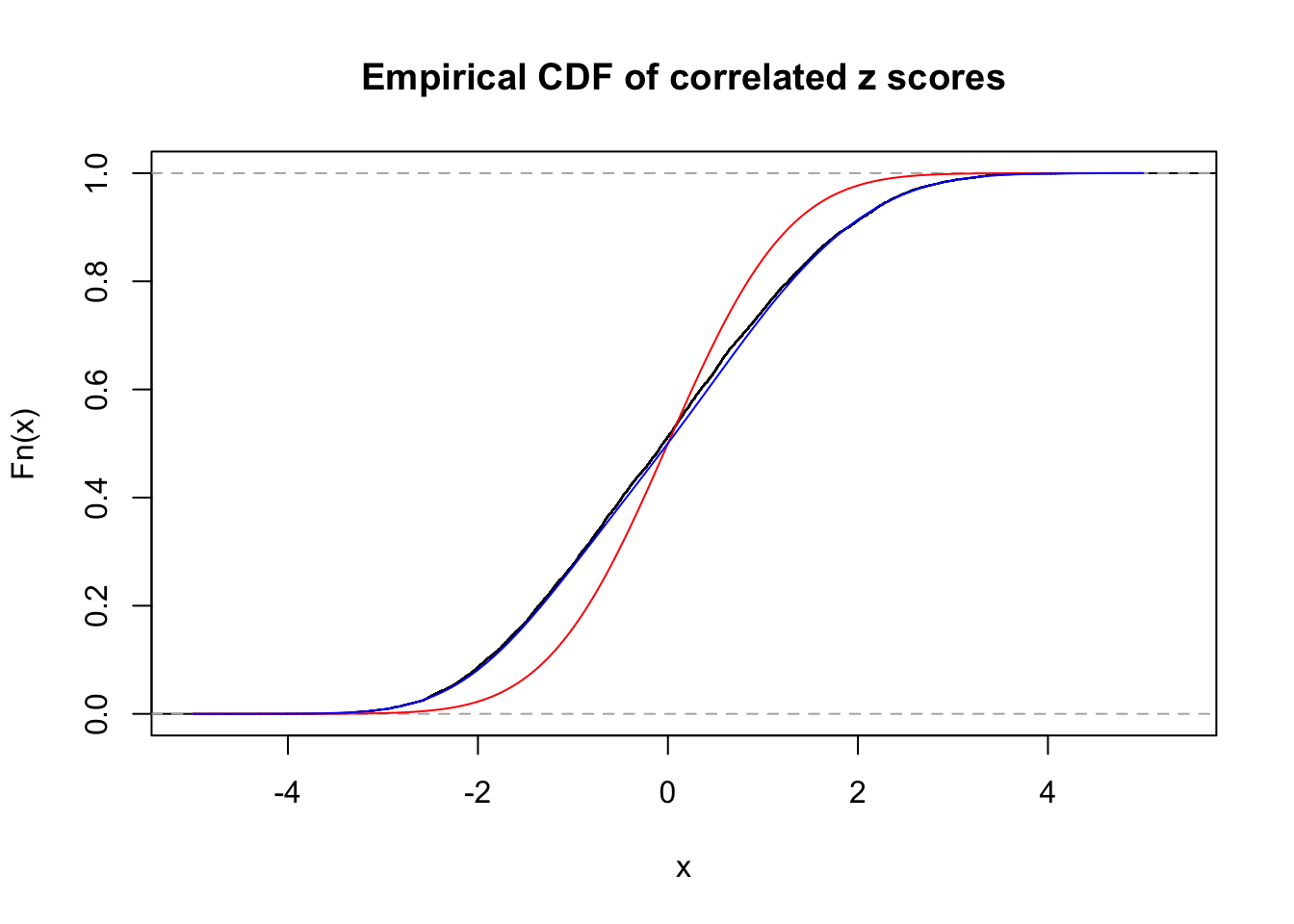
Data Set 771 :
Number of BH false discoveries: 12 ;
ASH's pihat0 = 0.04316169 ;
Normalized coefficients of Gaussian derivatives:
1 : 0 ; 2 : 0.73920141158121 ; 3 : 0.077182005444609 ; 4 : 0.167446690539398 ; 5 : 0.114289140310117 ; 6 : 0 ; 7 : 0.00550636320542386 ; 8 : 0.0464055959765927 ; 9 : 0 ; 10 : 0.0398398101639301 ;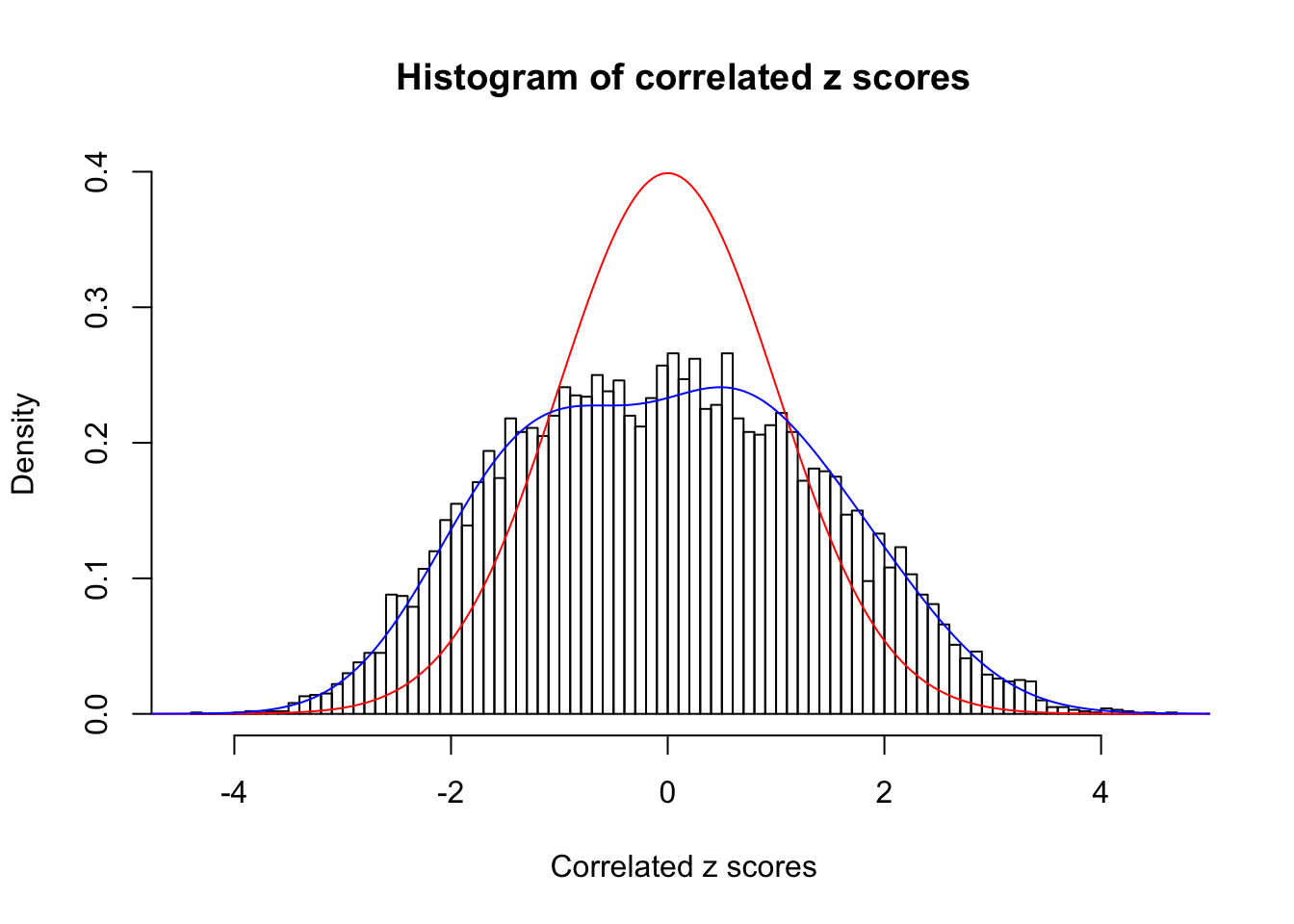
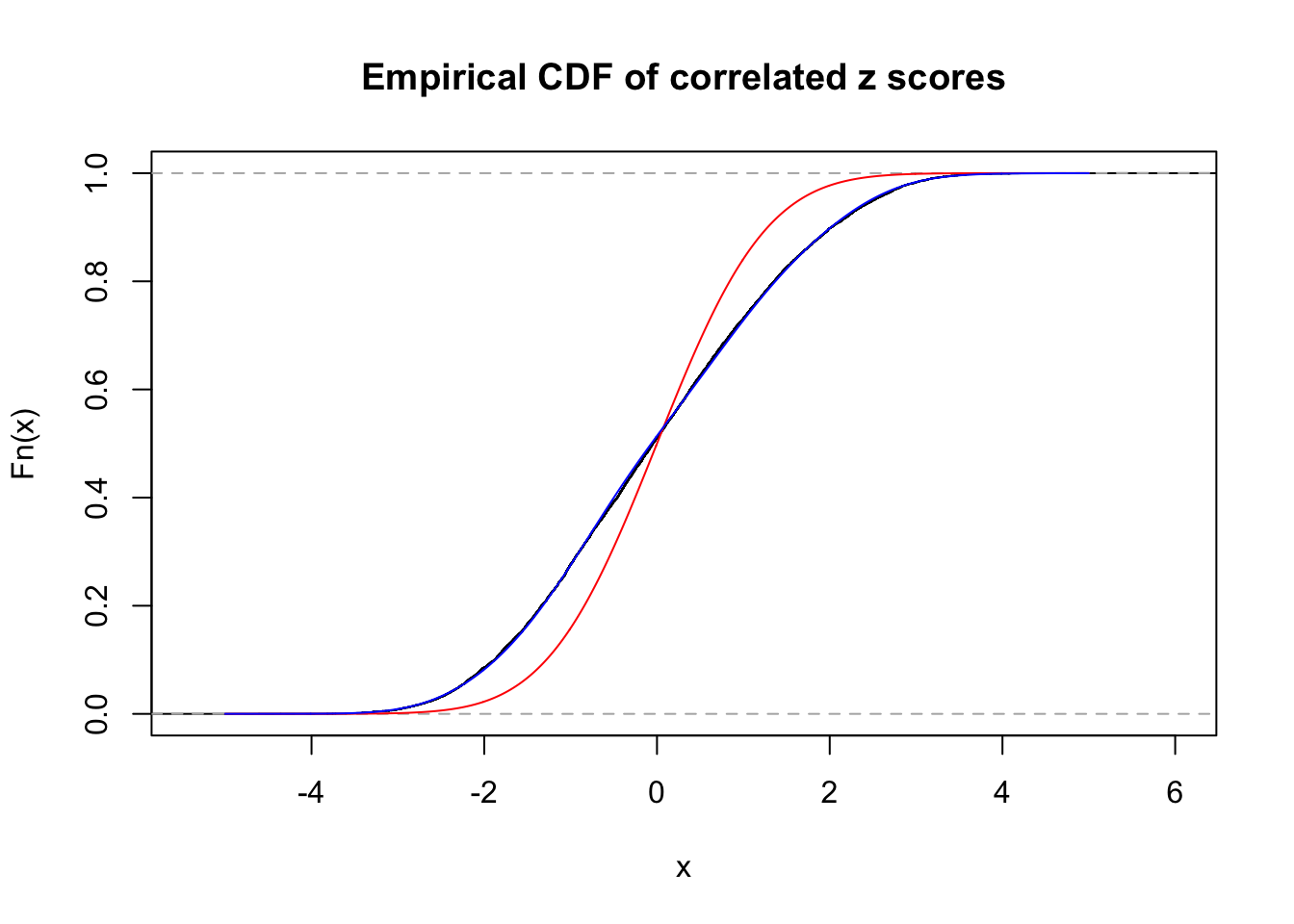
Data Set 389 :
Number of BH false discoveries: 11 ;
ASH's pihat0 = 0.03156173 ;
Normalized coefficients of Gaussian derivatives:
1 : 0.0104278129211743 ; 2 : 0.81975416226431 ; 3 : 0.175093847880514 ; 4 : 0.237691296370699 ; 5 : 0.0928656721296627 ; 6 : 0 ; 7 : 0 ; 8 : 0.0957748191090392 ; 9 : 0 ; 10 : 0.14869448917214 ;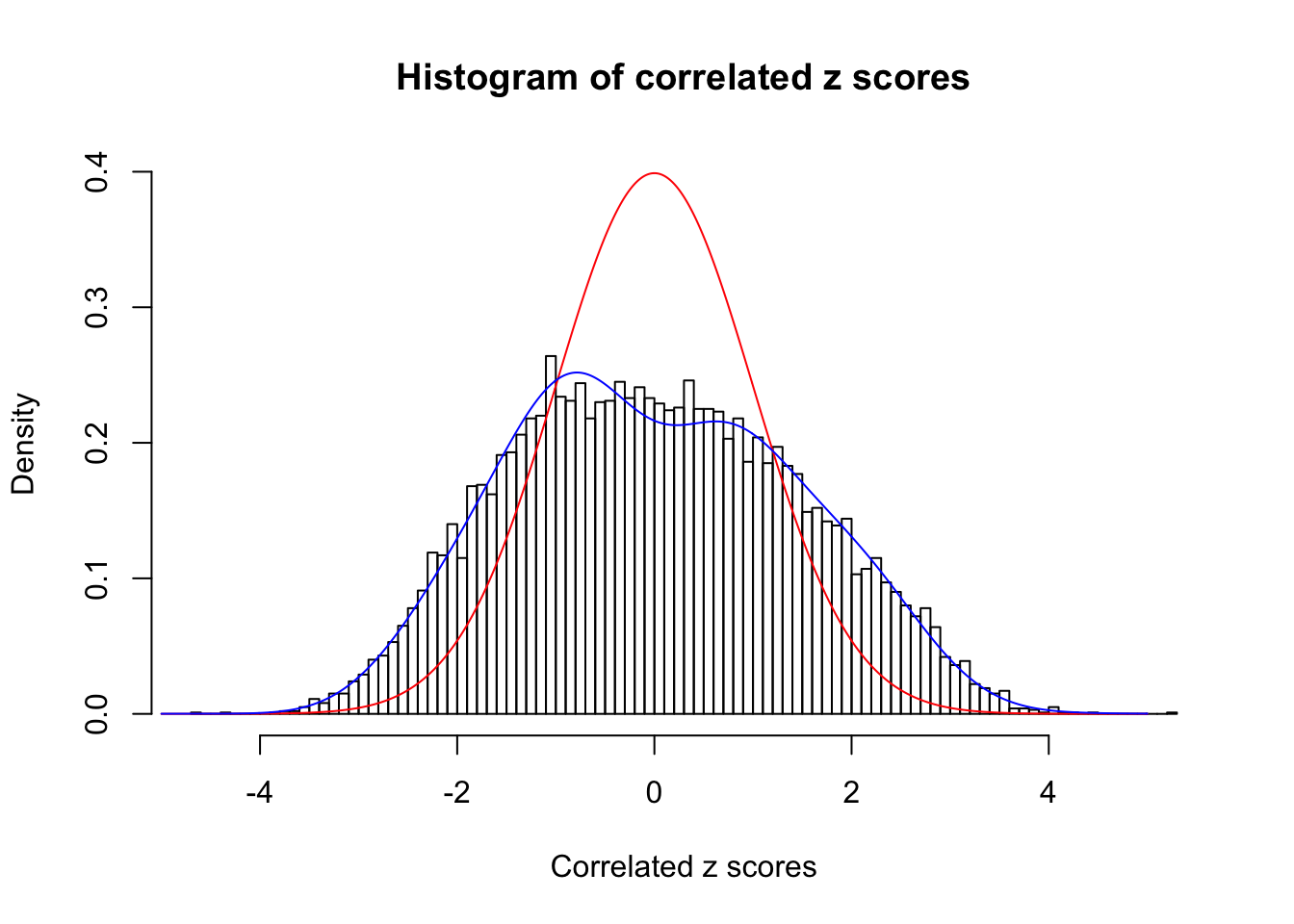
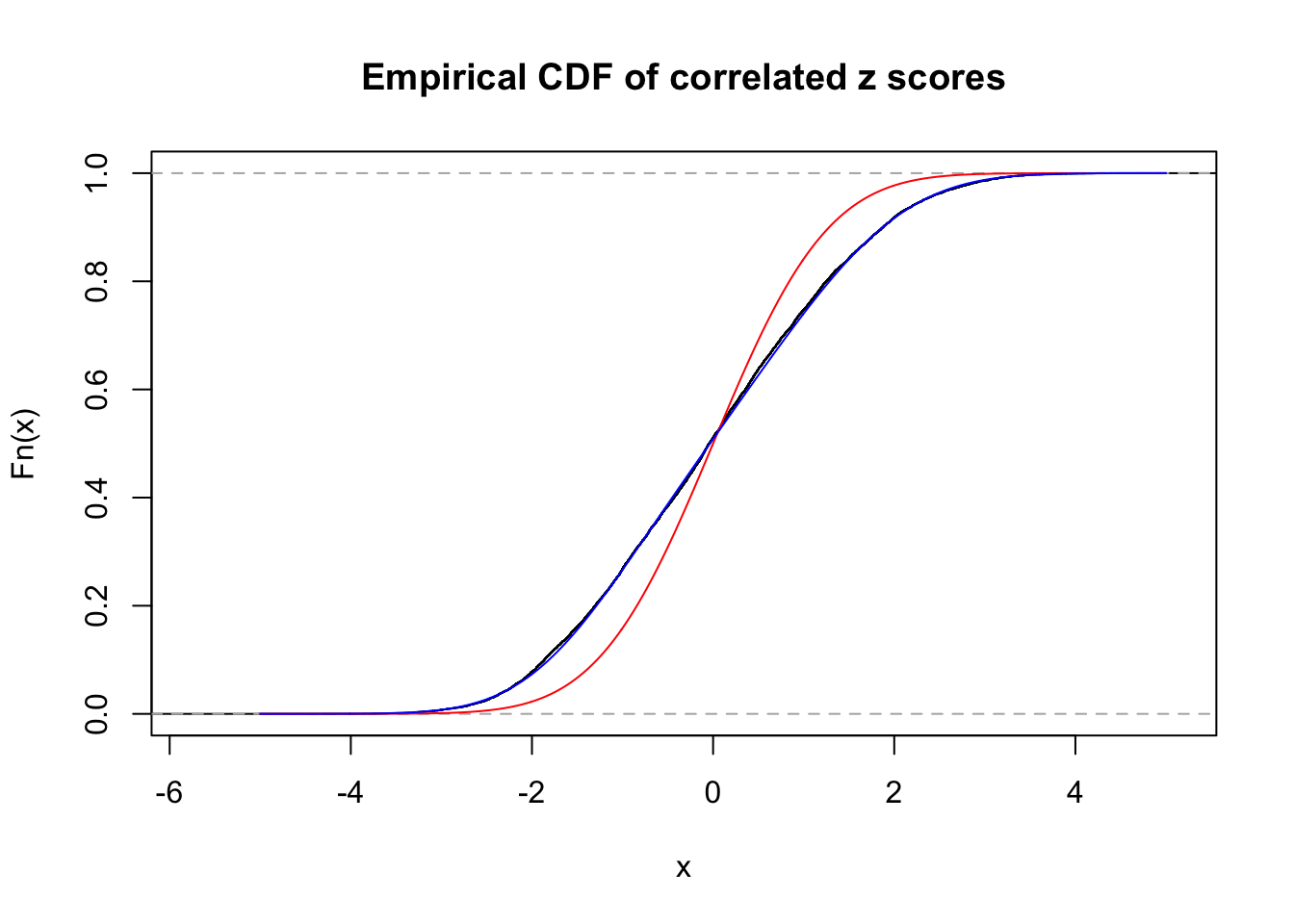
Data Set 485 :
Number of BH false discoveries: 9 ;
ASH's pihat0 = 0.04794909 ;
Normalized coefficients of Gaussian derivatives:
1 : 0 ; 2 : 0.688297974677999 ; 3 : 0.102717821163846 ; 4 : 0.1348683673964 ; 5 : 0.0774735270815003 ; 6 : 0 ; 7 : 0 ; 8 : 0 ; 9 : 0 ; 10 : 0 ;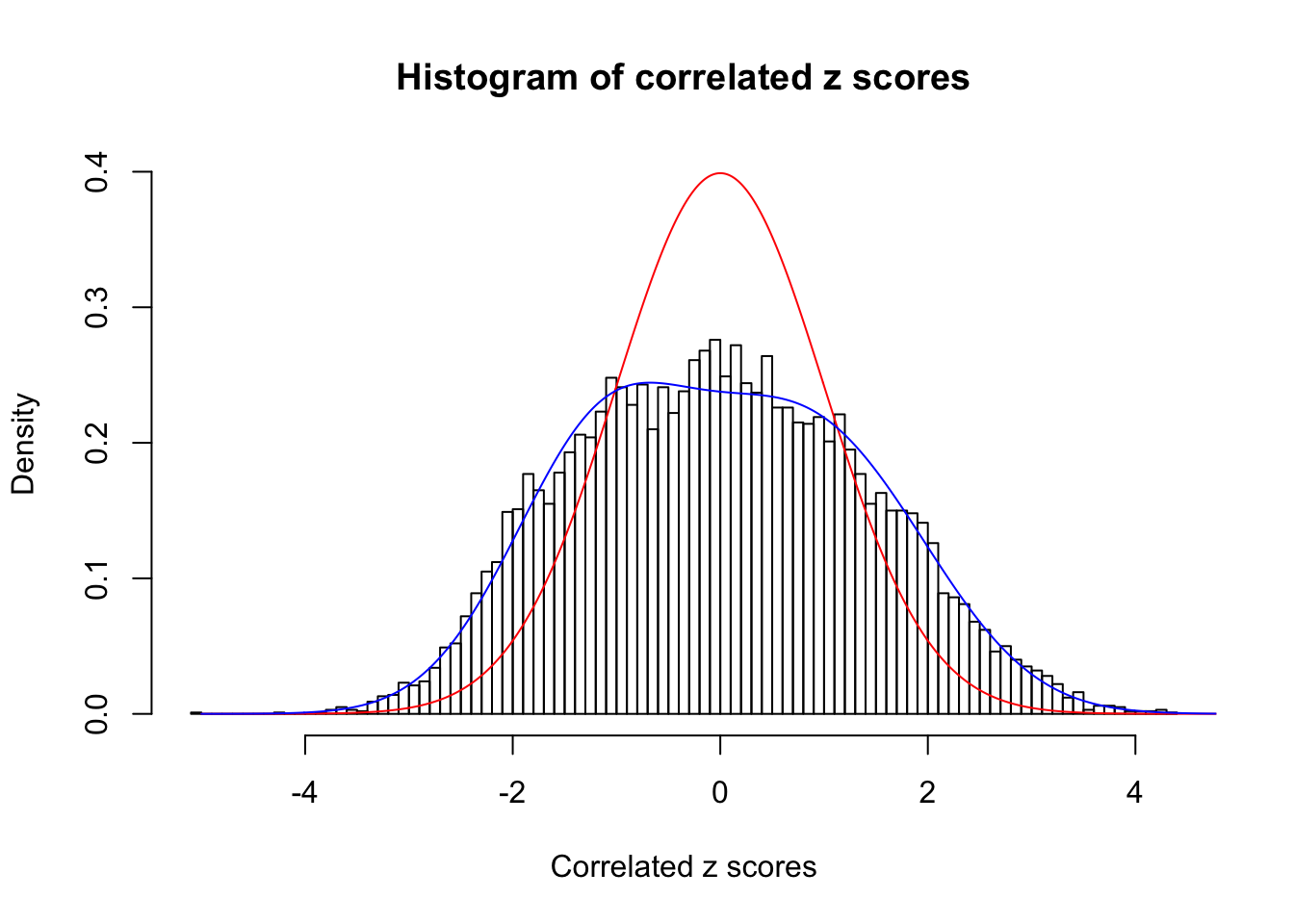
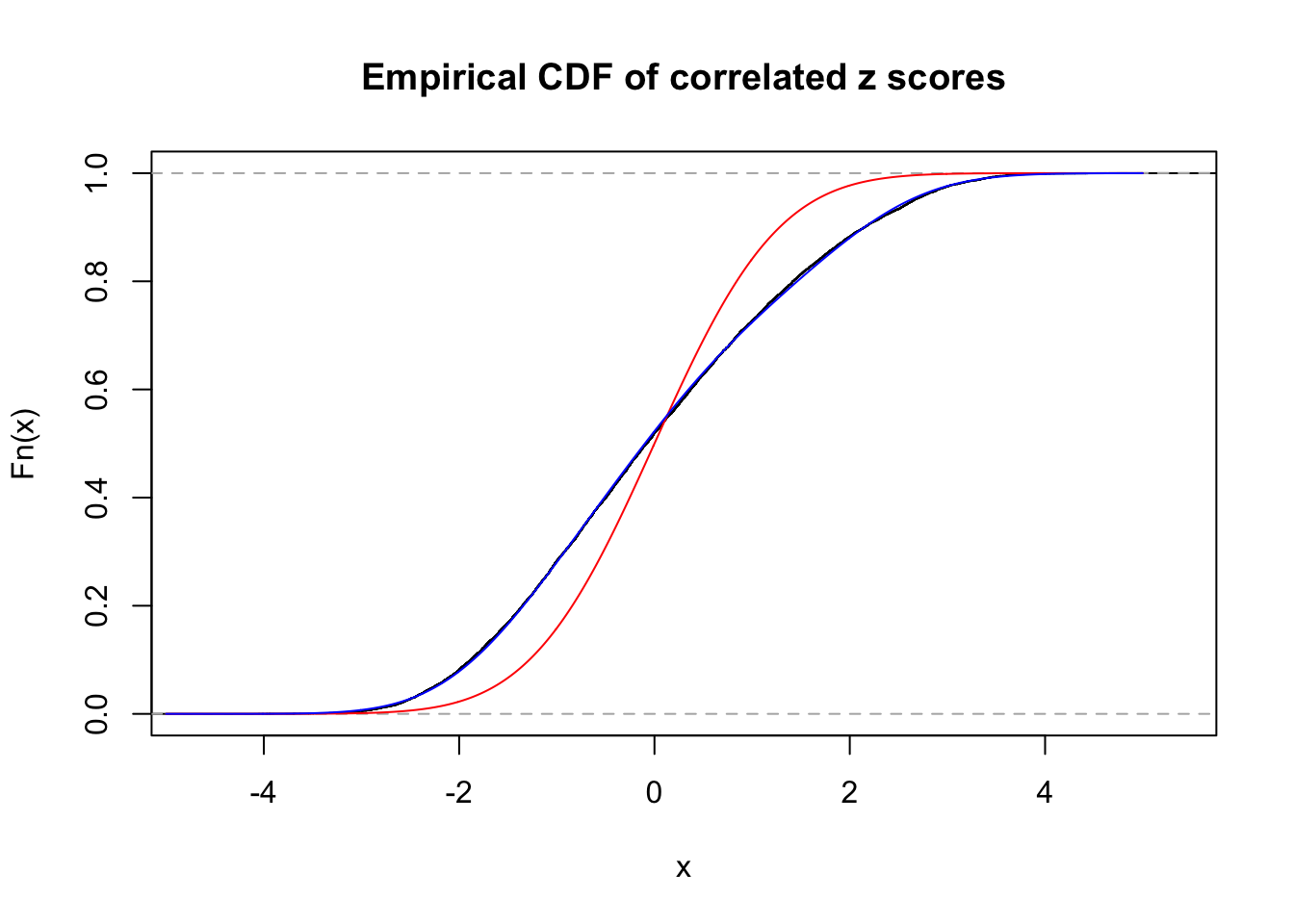
Data Set 77 :
Number of BH false discoveries: 7 ;
ASH's pihat0 = 0.05151585 ;
Normalized coefficients of Gaussian derivatives:
1 : 0.0316698203536559 ; 2 : 0.900083565443527 ; 3 : 0.369105500650609 ; 4 : 0.348606888712178 ; 5 : 0.225564147471124 ; 6 : 0 ; 7 : 0 ; 8 : 0 ; 9 : 0 ; 10 : 0 ;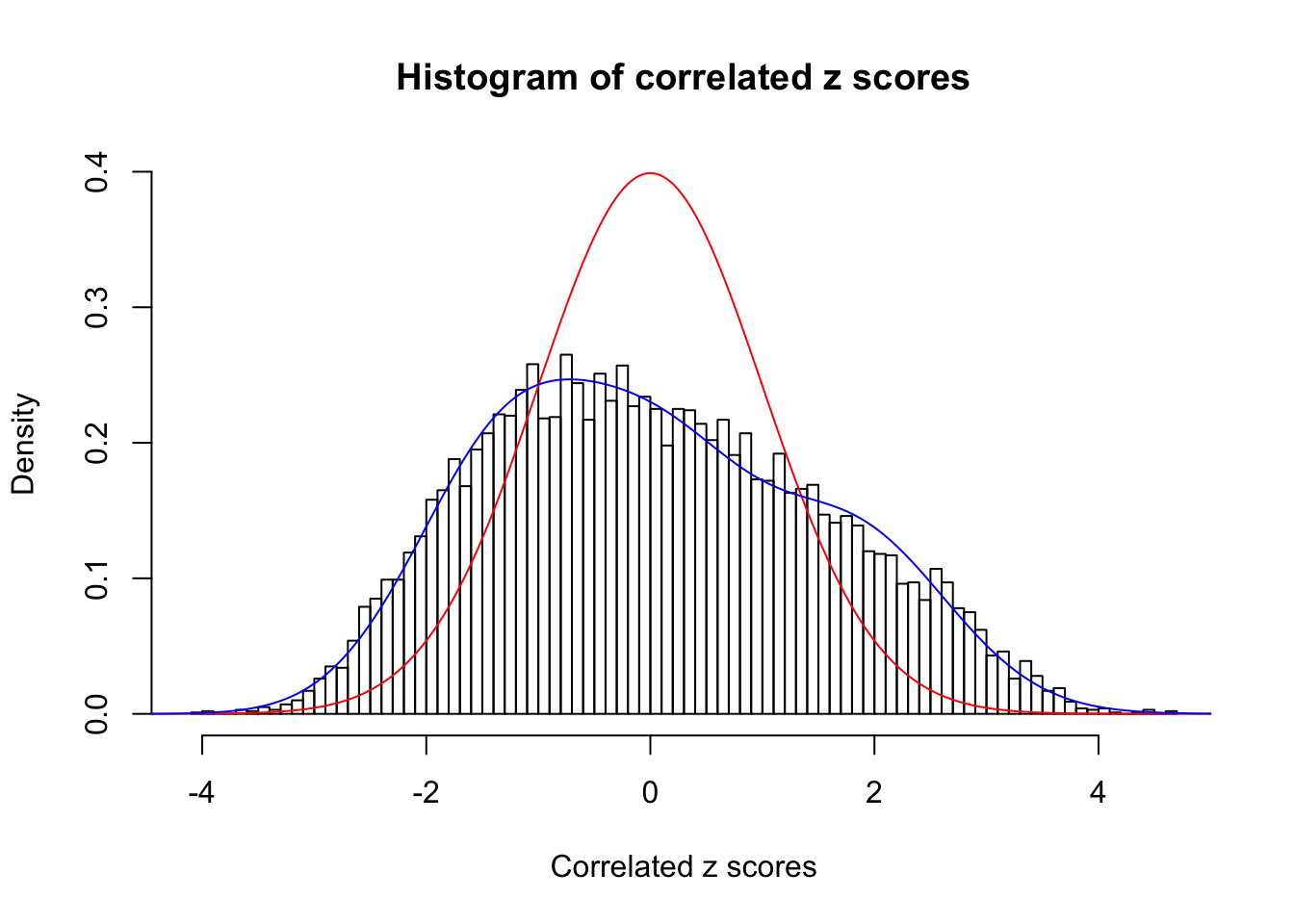
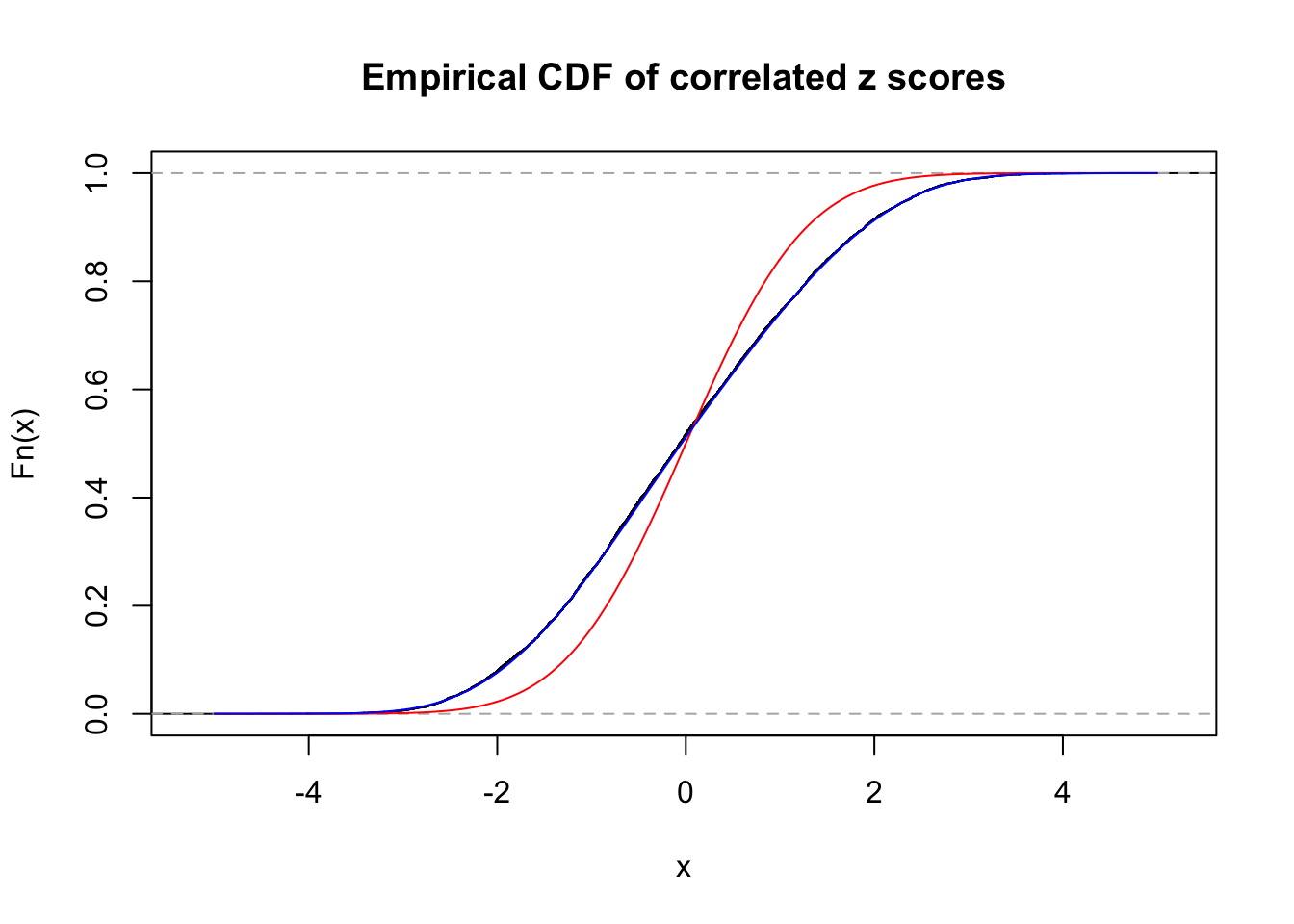
Data Set 984 :
Number of BH false discoveries: 7 ;
ASH's pihat0 = 0.04567195 ;
Normalized coefficients of Gaussian derivatives:
1 : 0 ; 2 : 0.699043214591173 ; 3 : 0.100302581737407 ; 4 : 0.145190970625763 ; 5 : 0.0474679405200551 ; 6 : 0 ; 7 : 0 ; 8 : 0.134635889035747 ; 9 : 0 ; 10 : 0.110318005619196 ;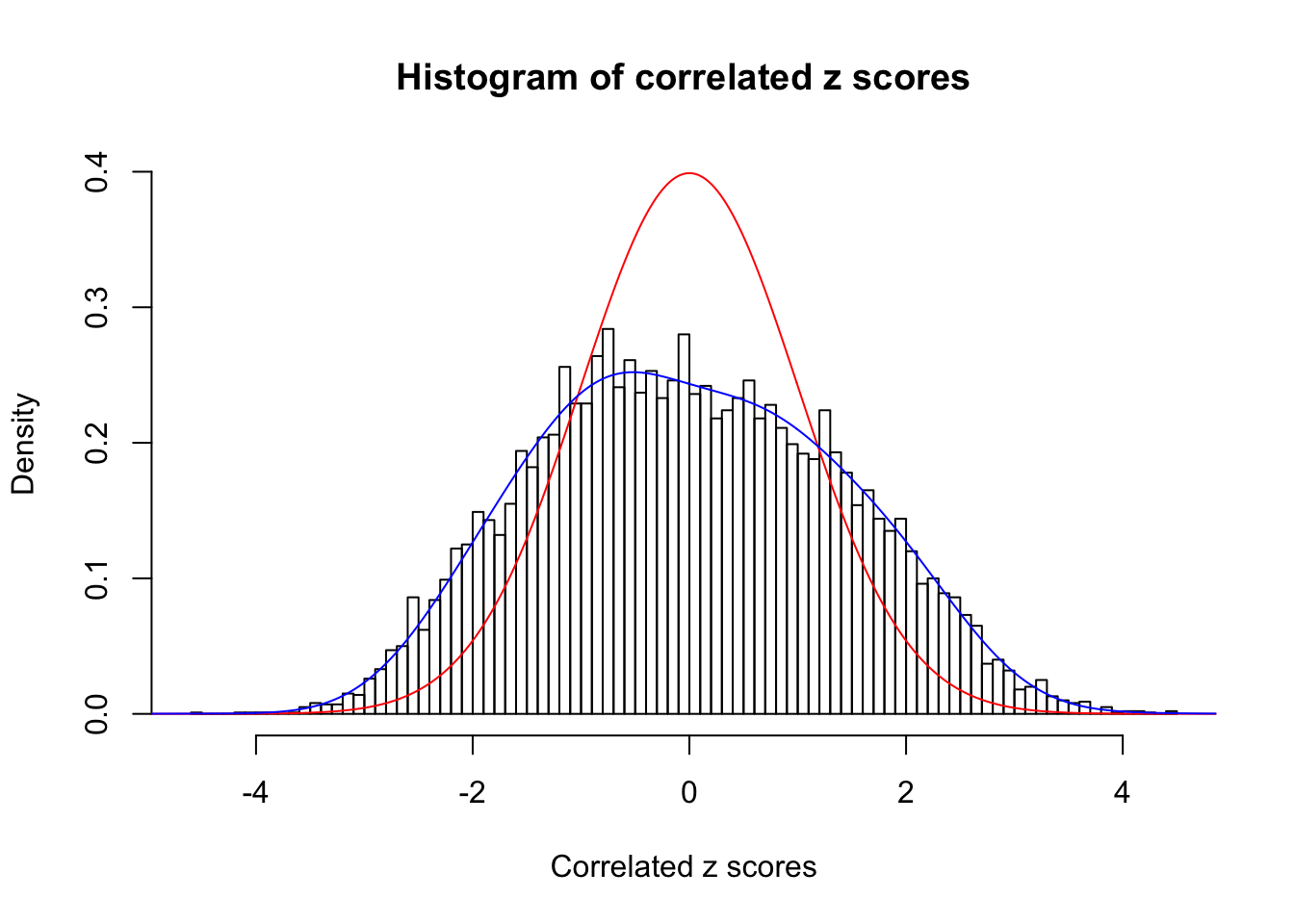
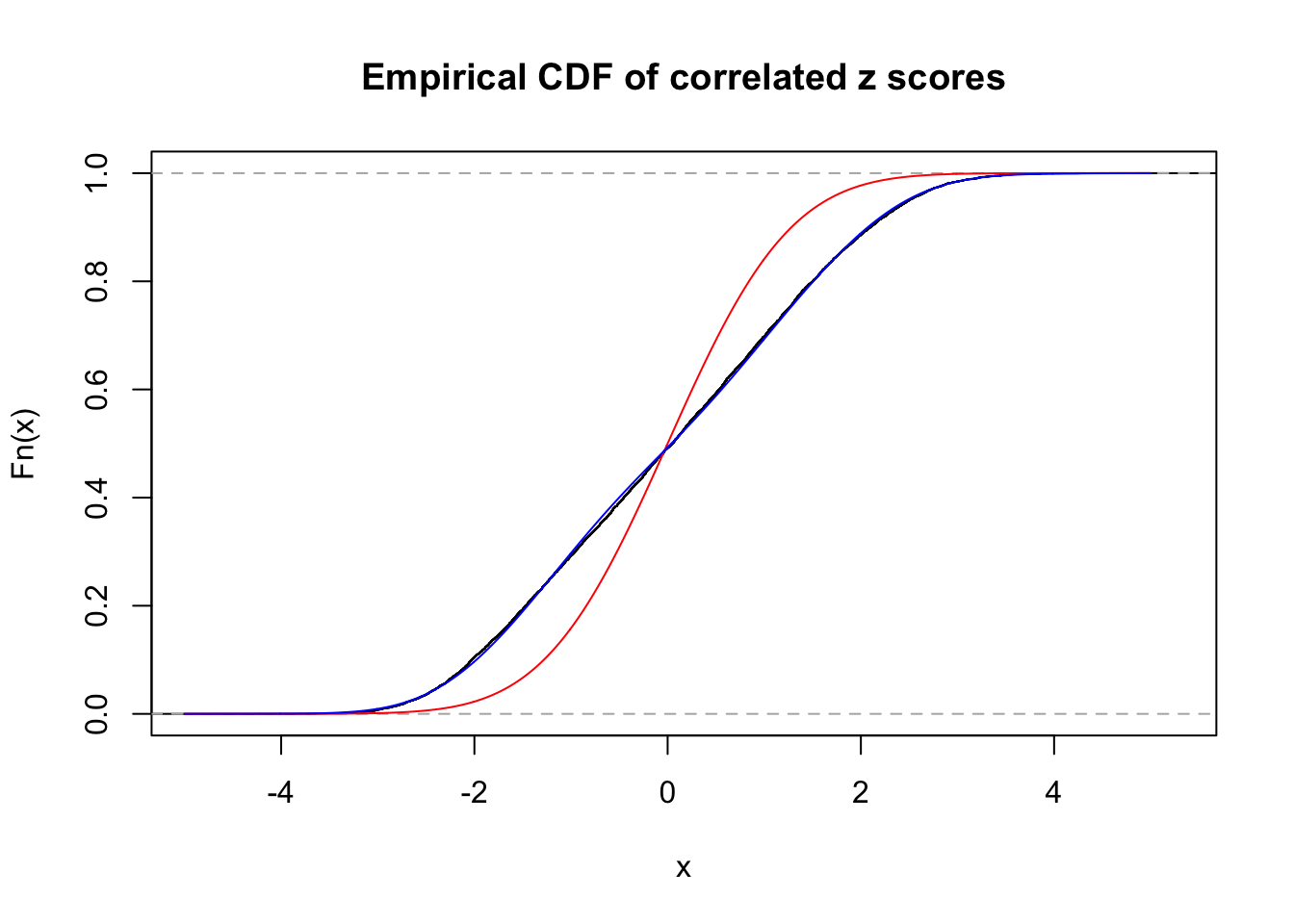
Data Set 360 :
Number of BH false discoveries: 6 ;
ASH's pihat0 = 0.0308234 ;
Normalized coefficients of Gaussian derivatives:
1 : 0.0370729985554797 ; 2 : 0.965843584122268 ; 3 : 0.113777617637278 ; 4 : 0.182892304104783 ; 5 : 0.0985758556513691 ; 6 : 0 ; 7 : 0 ; 8 : 0.175602983077514 ; 9 : 0 ; 10 : 0.128160801611689 ;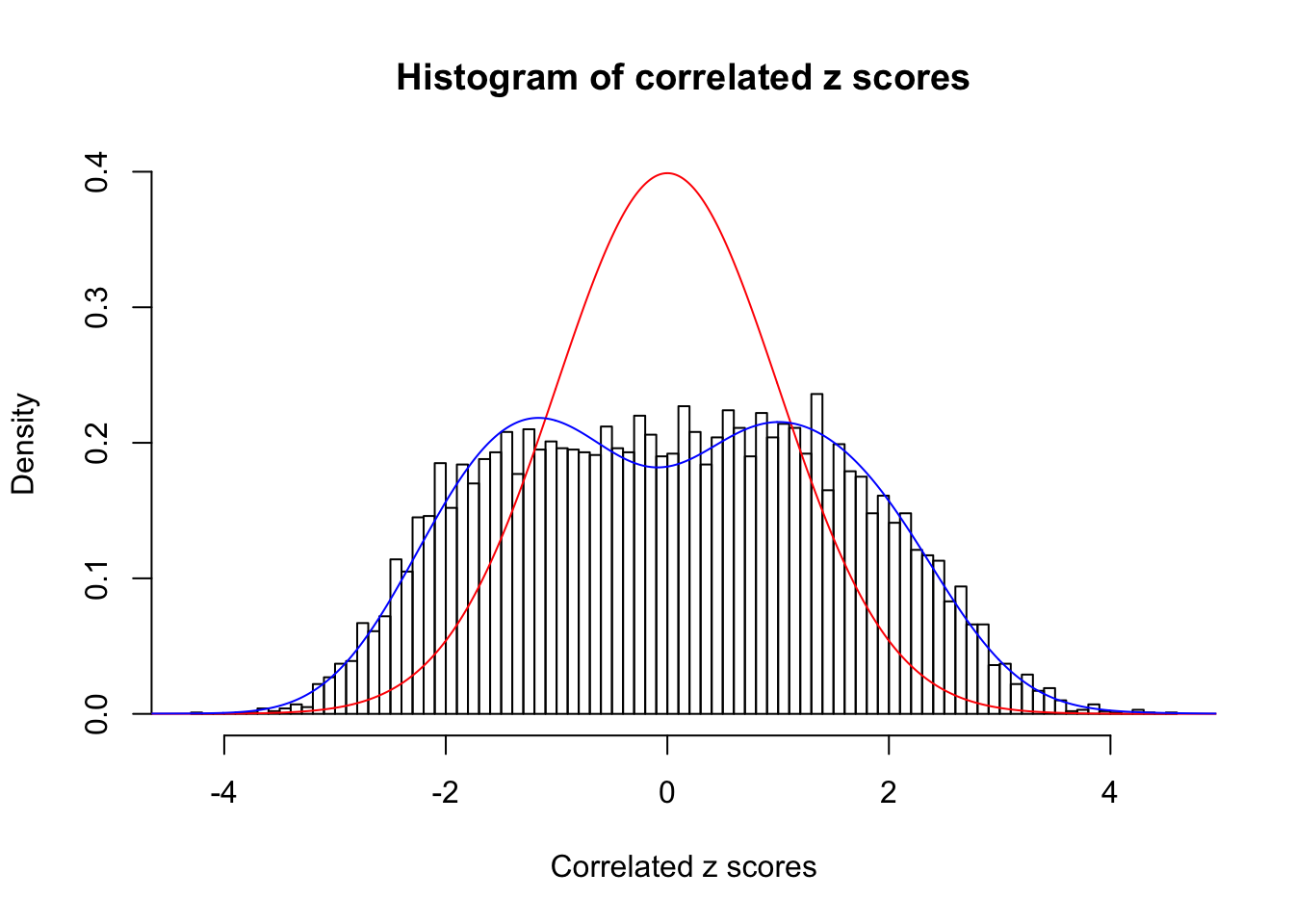
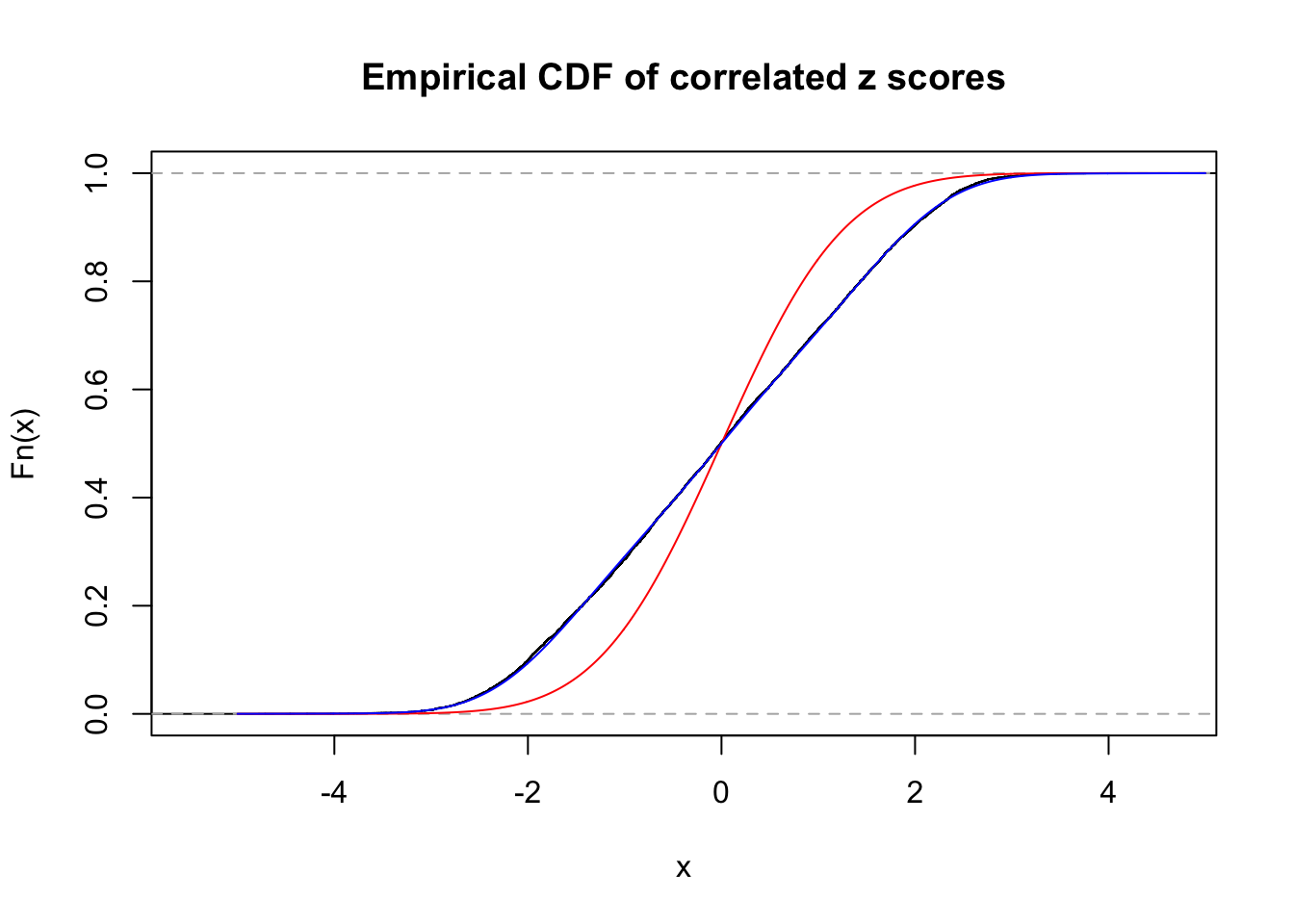
Data Set 522 :
Number of BH false discoveries: 4 ;
ASH's pihat0 = 0.02083846 ;
Normalized coefficients of Gaussian derivatives:
1 : 0 ; 2 : 0.836683104968565 ; 3 : 0 ; 4 : 0.0380324164945556 ; 5 : 0 ; 6 : 0 ; 7 : 0 ; 8 : 0.299307885307397 ; 9 : 0 ; 10 : 0.0990067134691387 ;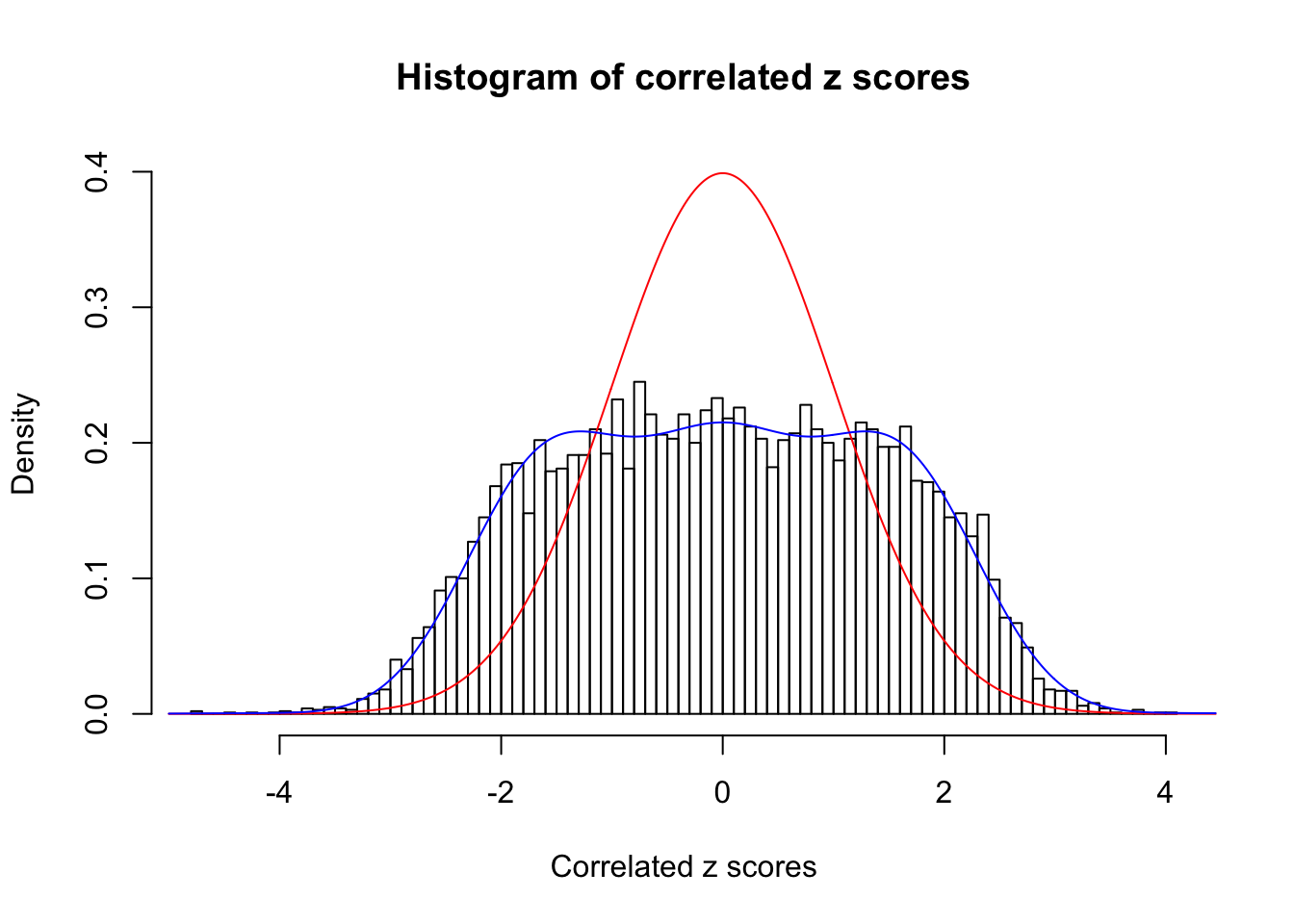
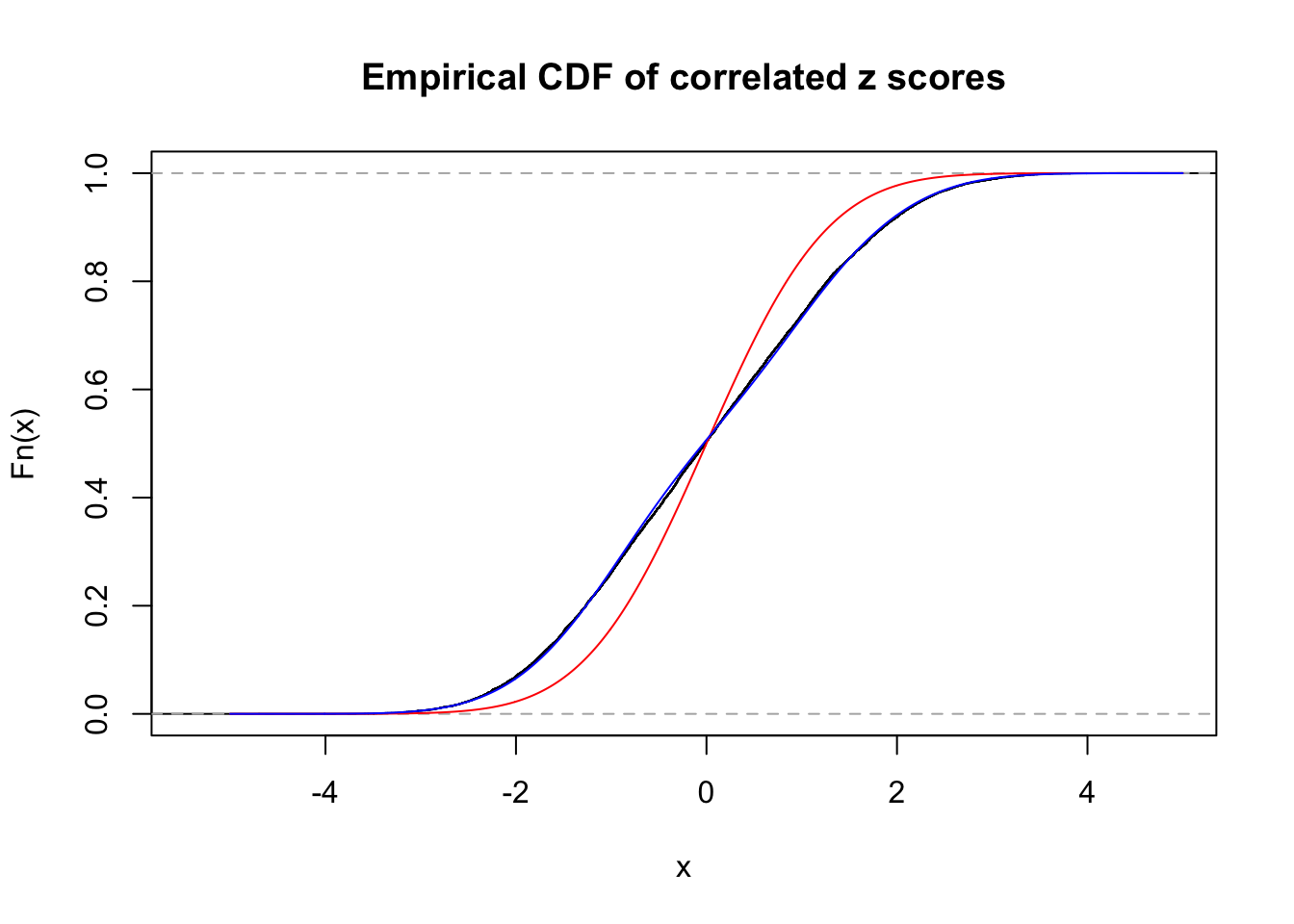
Data Set 22 :
Number of BH false discoveries: 1 ;
ASH's pihat0 = 0.03271534 ;
Normalized coefficients of Gaussian derivatives:
1 : 0.011182140702874 ; 2 : 0.637669154664922 ; 3 : 0.0954740117610049 ; 4 : 0.0145362179577236 ; 5 : 0.0384875211132818 ; 6 : 0 ; 7 : 0 ; 8 : 0 ; 9 : 0 ; 10 : 0.0366040528004269 ;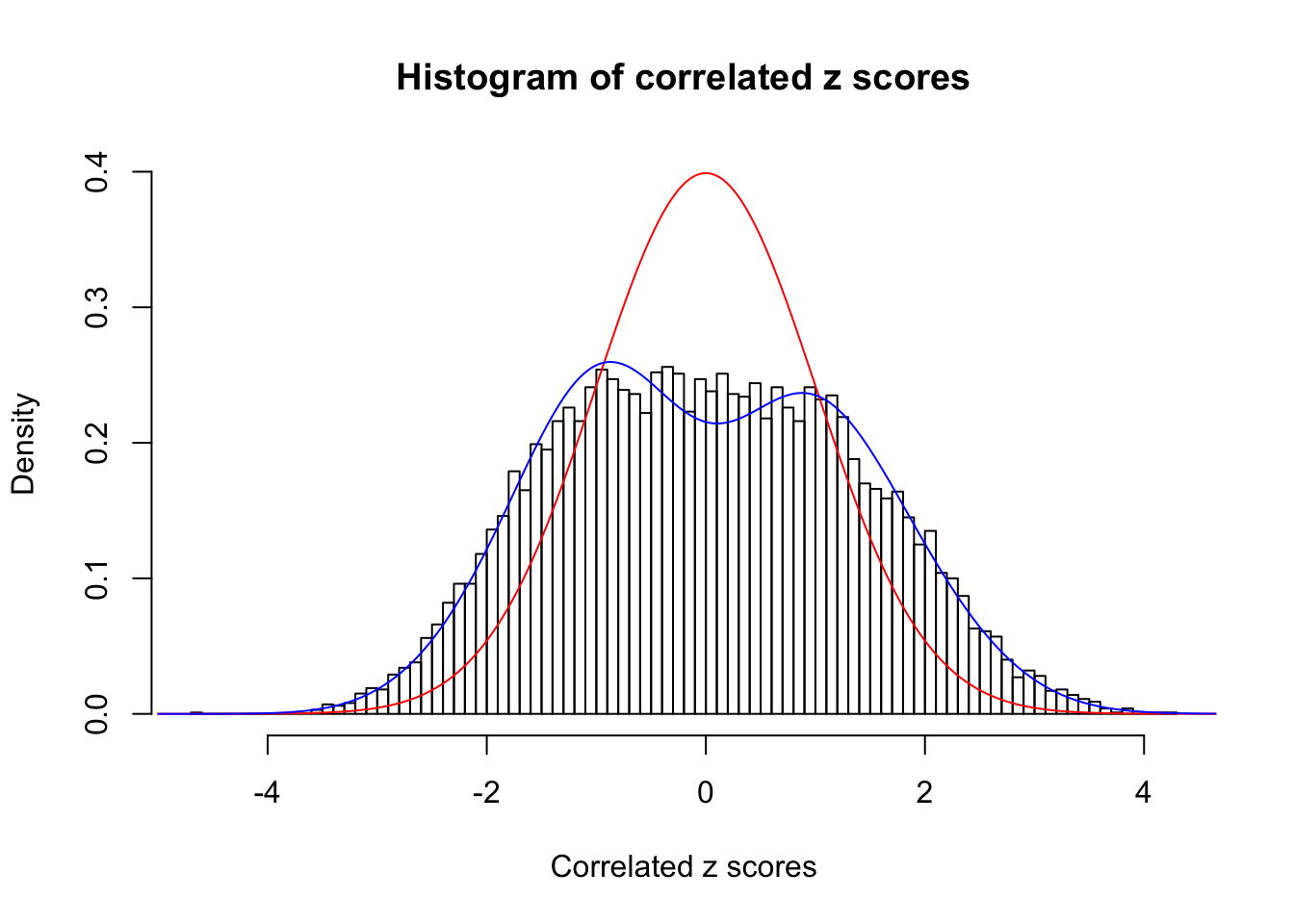
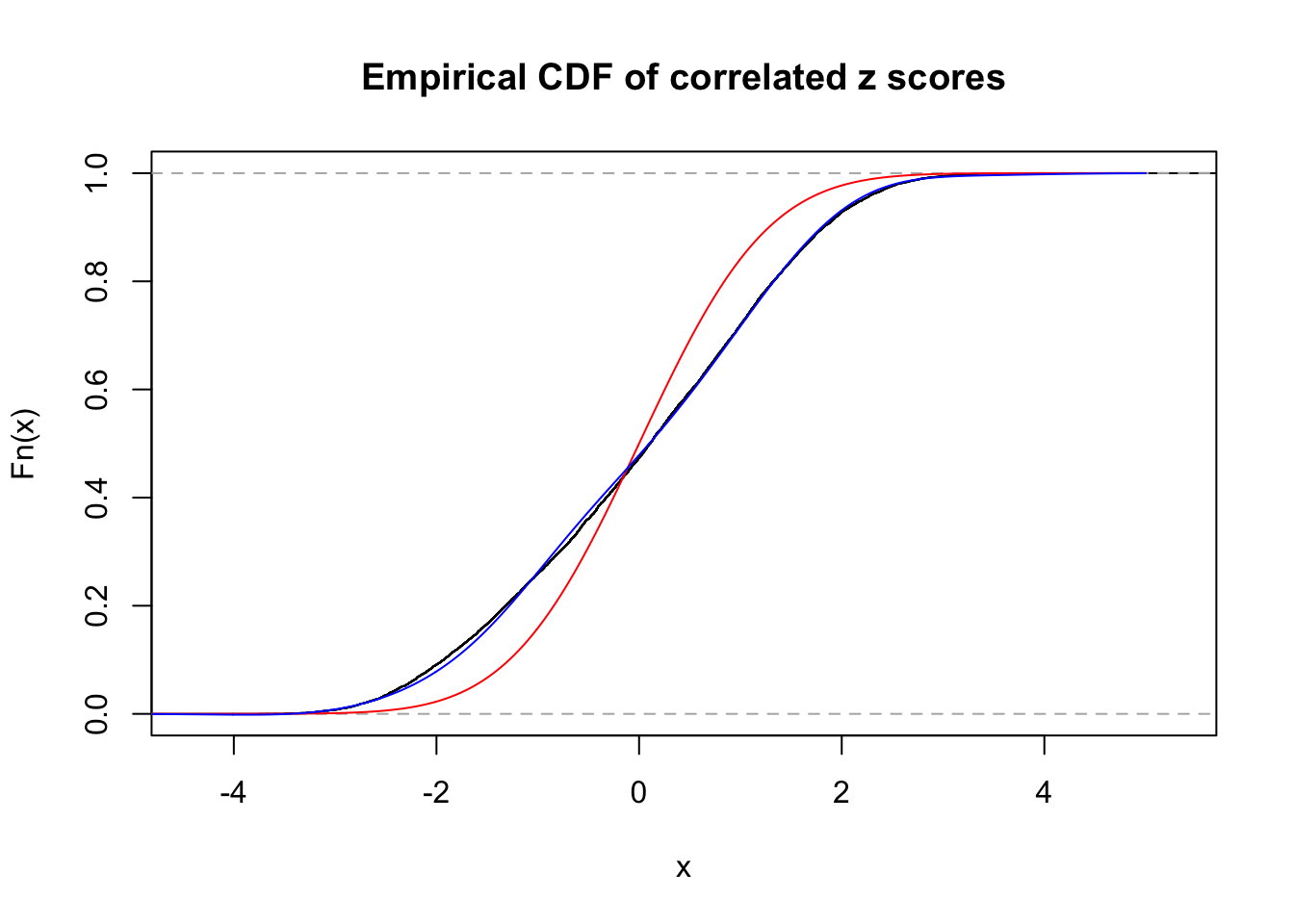
Data Set 269 :
Number of BH false discoveries: 1 ;
ASH's pihat0 = 0.02787007 ;
Normalized coefficients of Gaussian derivatives:
1 : 0.0350127752264117 ; 2 : 0.665763246213506 ; 3 : 0 ; 4 : 0 ; 5 : 0.360745065018111 ; 6 : 0 ; 7 : 0.660778015759139 ; 8 : 0 ; 9 : 0.341451404165077 ; 10 : 0 ;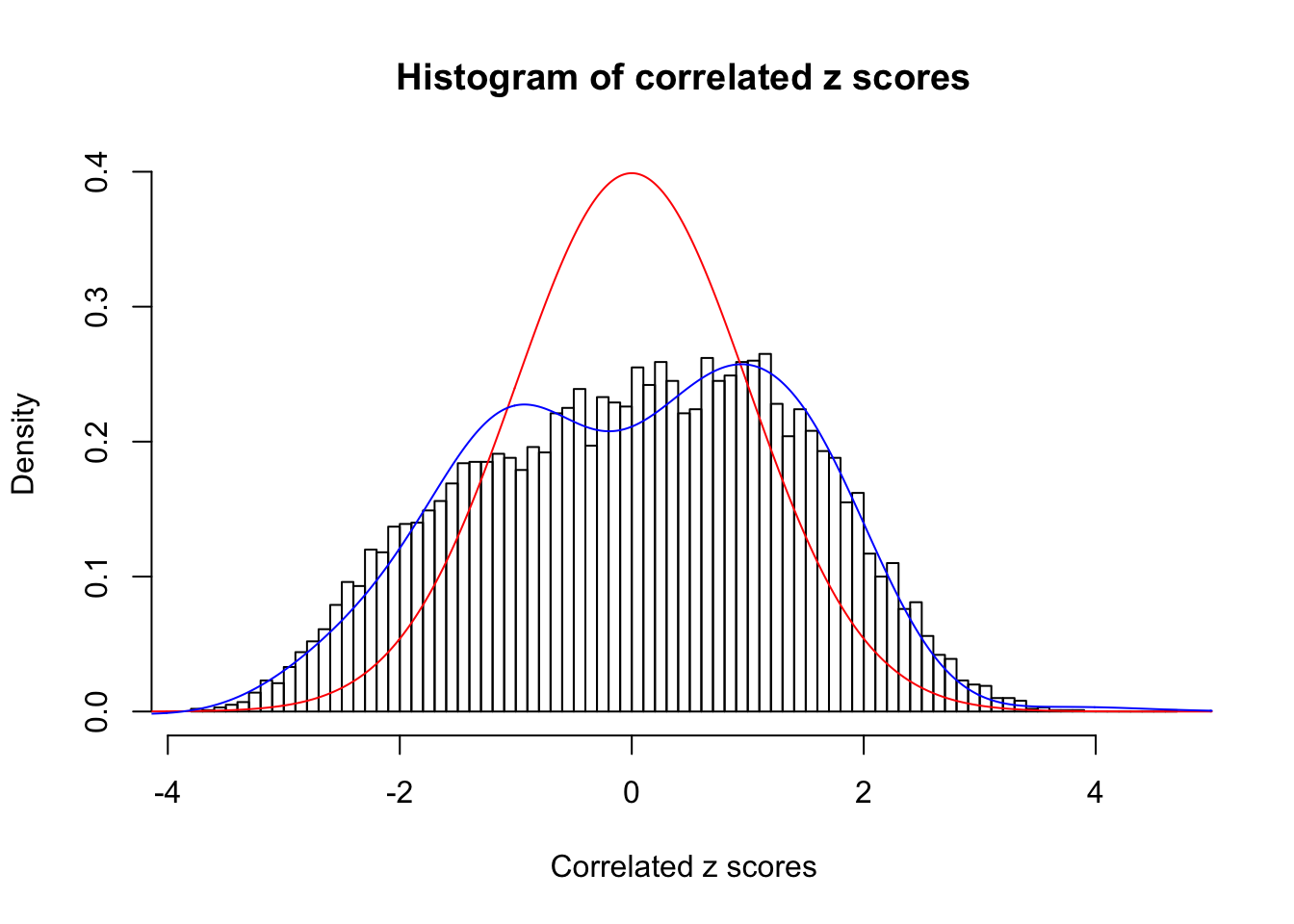
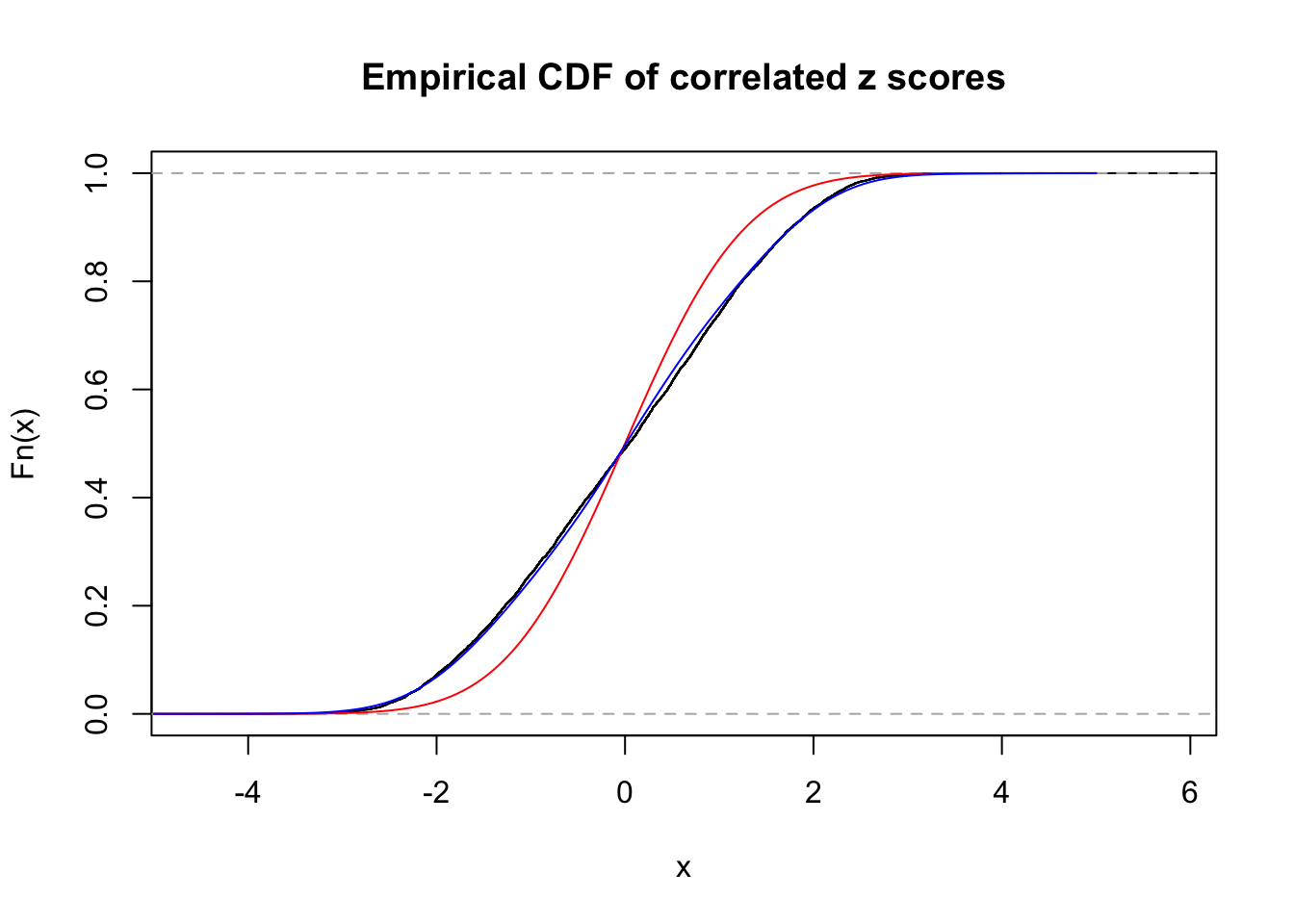
Data Set 403 :
Number of BH false discoveries: 1 ;
ASH's pihat0 = 0.0531628 ;
Normalized coefficients of Gaussian derivatives:
1 : 0.00287620650301181 ; 2 : 0.547191548824974 ; 3 : 0 ; 4 : 0 ; 5 : 0.0381682188675178 ; 6 : 0.000237362926646135 ; 7 : 0.0760788952836512 ; 8 : 0.155507732499129 ; 9 : 0.0557968950438762 ; 10 : 0.00176813778552284 ;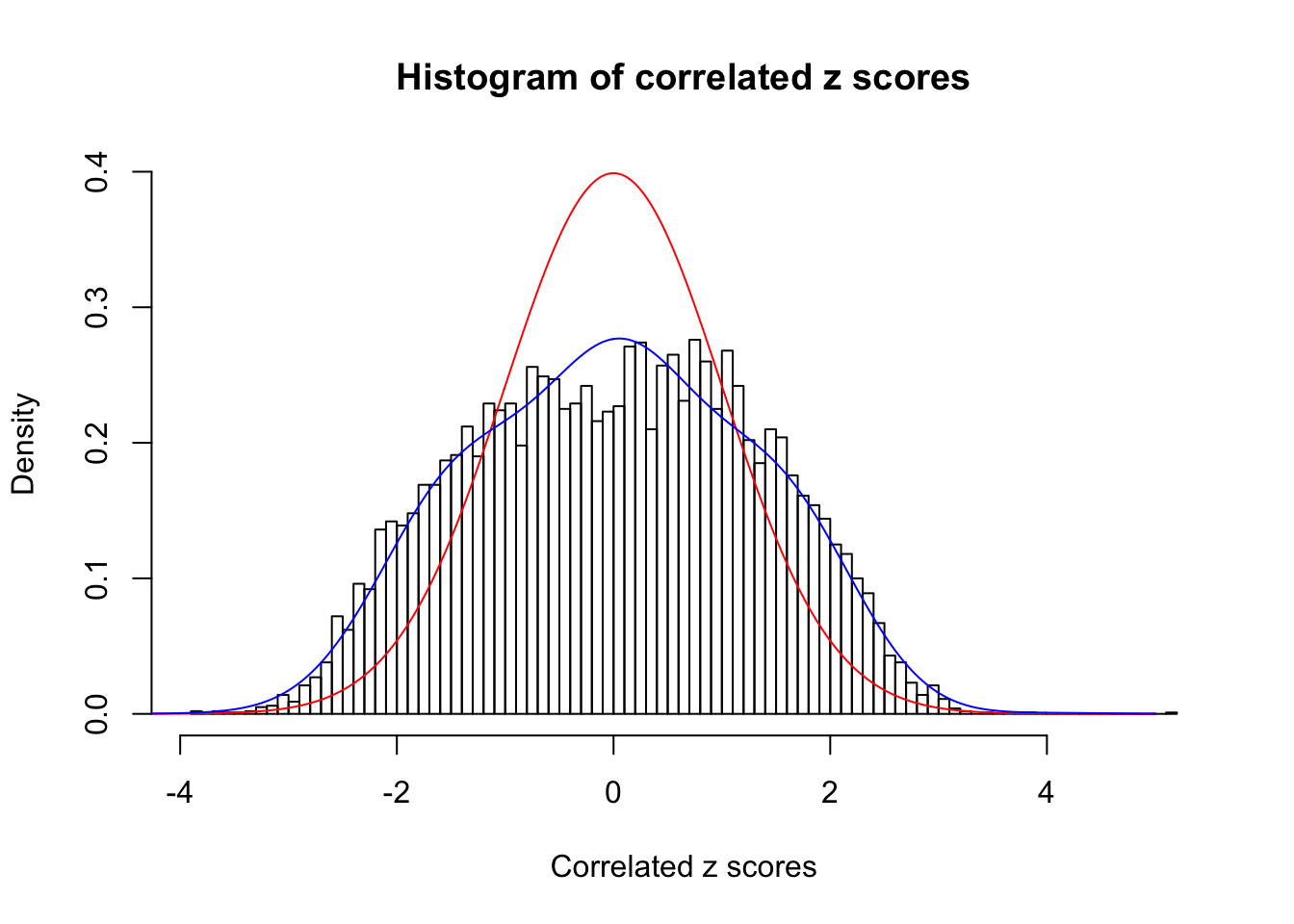
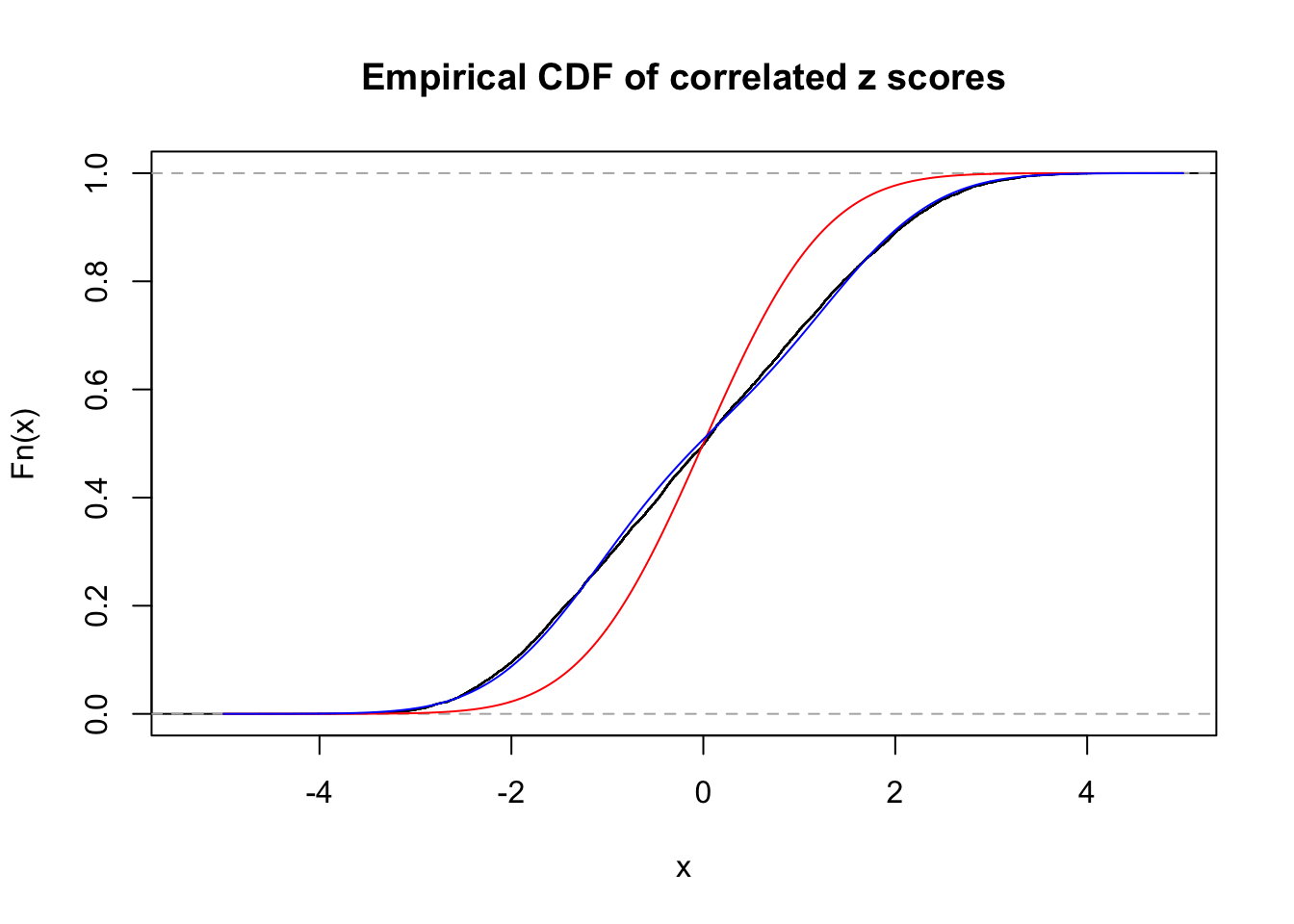
Data Set 923 :
Number of BH false discoveries: 1 ;
ASH's pihat0 = 0.03239883 ;
Normalized coefficients of Gaussian derivatives:
1 : 0.0237509340668928 ; 2 : 0.920816163763449 ; 3 : 0.126877447333694 ; 4 : 0.158793996465808 ; 5 : 0.0298935777348498 ; 6 : 0 ; 7 : 0 ; 8 : 0 ; 9 : 0 ; 10 : 0 ;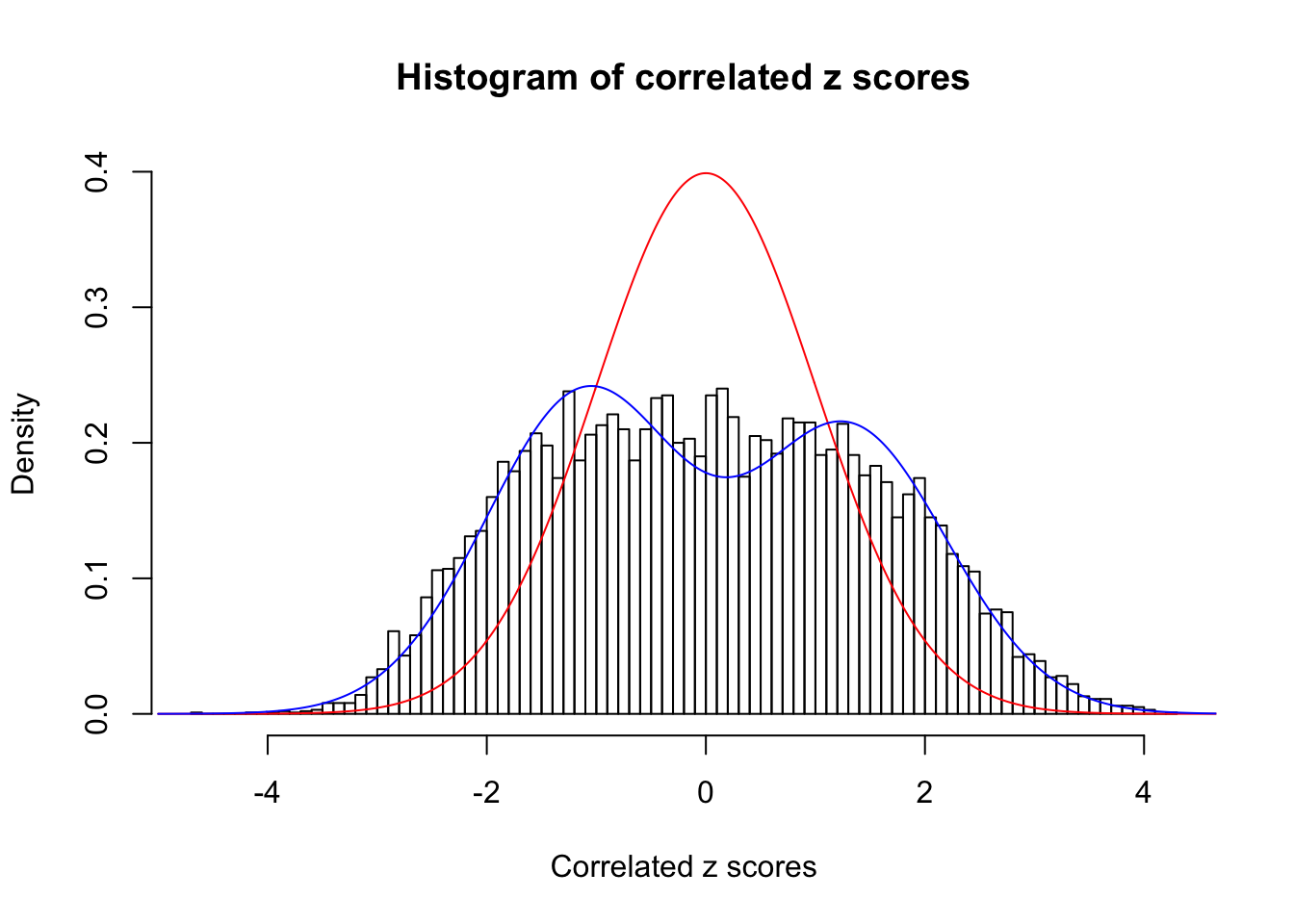
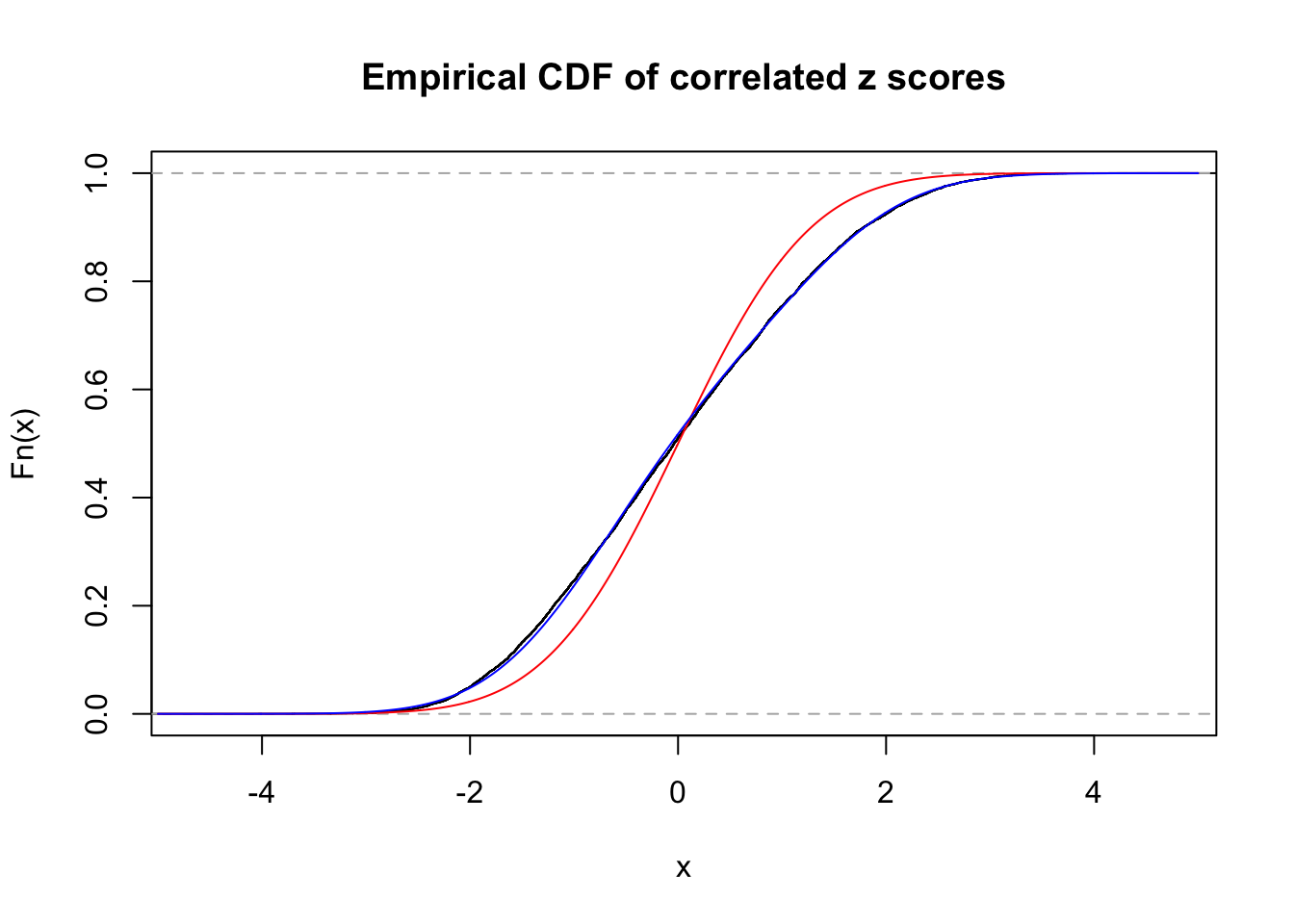
Data Set 3 :
Number of BH false discoveries: 0 ;
ASH's pihat0 = 0.0465218 ;
Normalized coefficients of Gaussian derivatives:
1 : 0.0232314826078553 ; 2 : 0.491281828479503 ; 3 : 0.165633823951823 ; 4 : 0 ; 5 : 0 ; 6 : 0 ; 7 : 0 ; 8 : 0 ; 9 : 0 ; 10 : 0 ;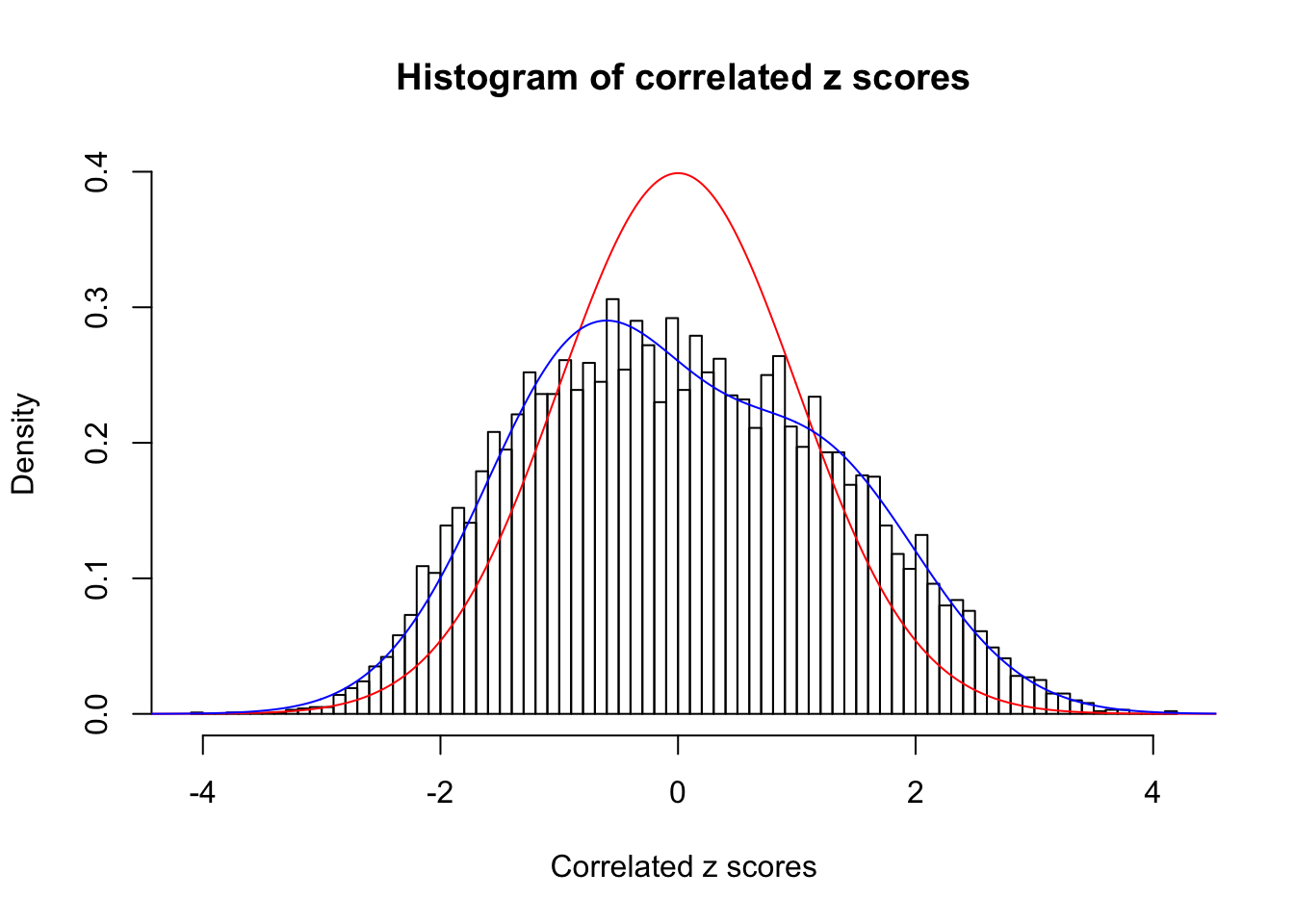
Good
These data sets are friendly for both BH and ASH.
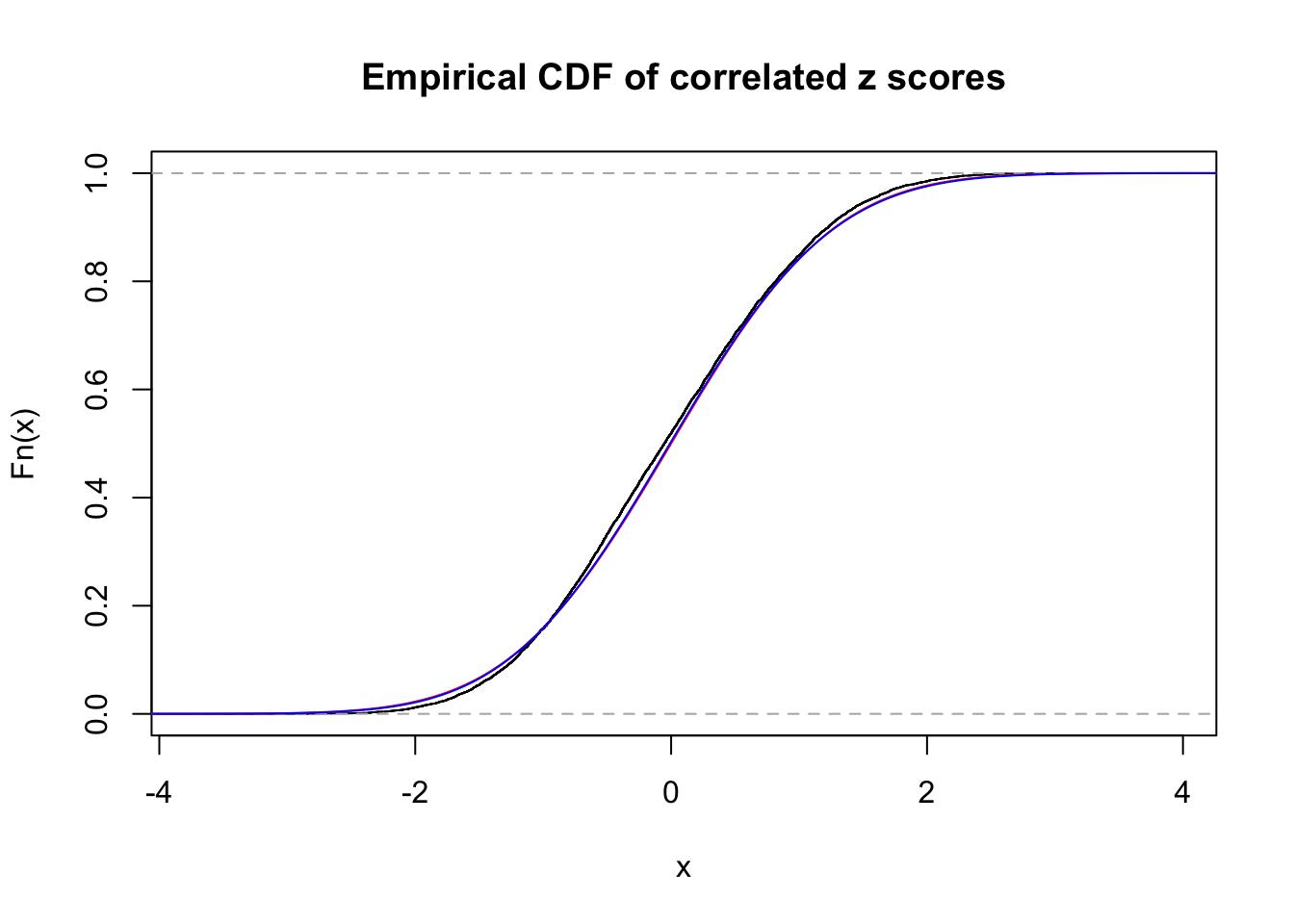
Data Set 4 :
Number of BH false discoveries: 0 ;
ASH's pihat0 = 1 ;
Normalized coefficients of Gaussian derivatives:
1 : 0 ; 2 : 0 ; 3 : 0.0220506376197135 ; 4 : 0 ;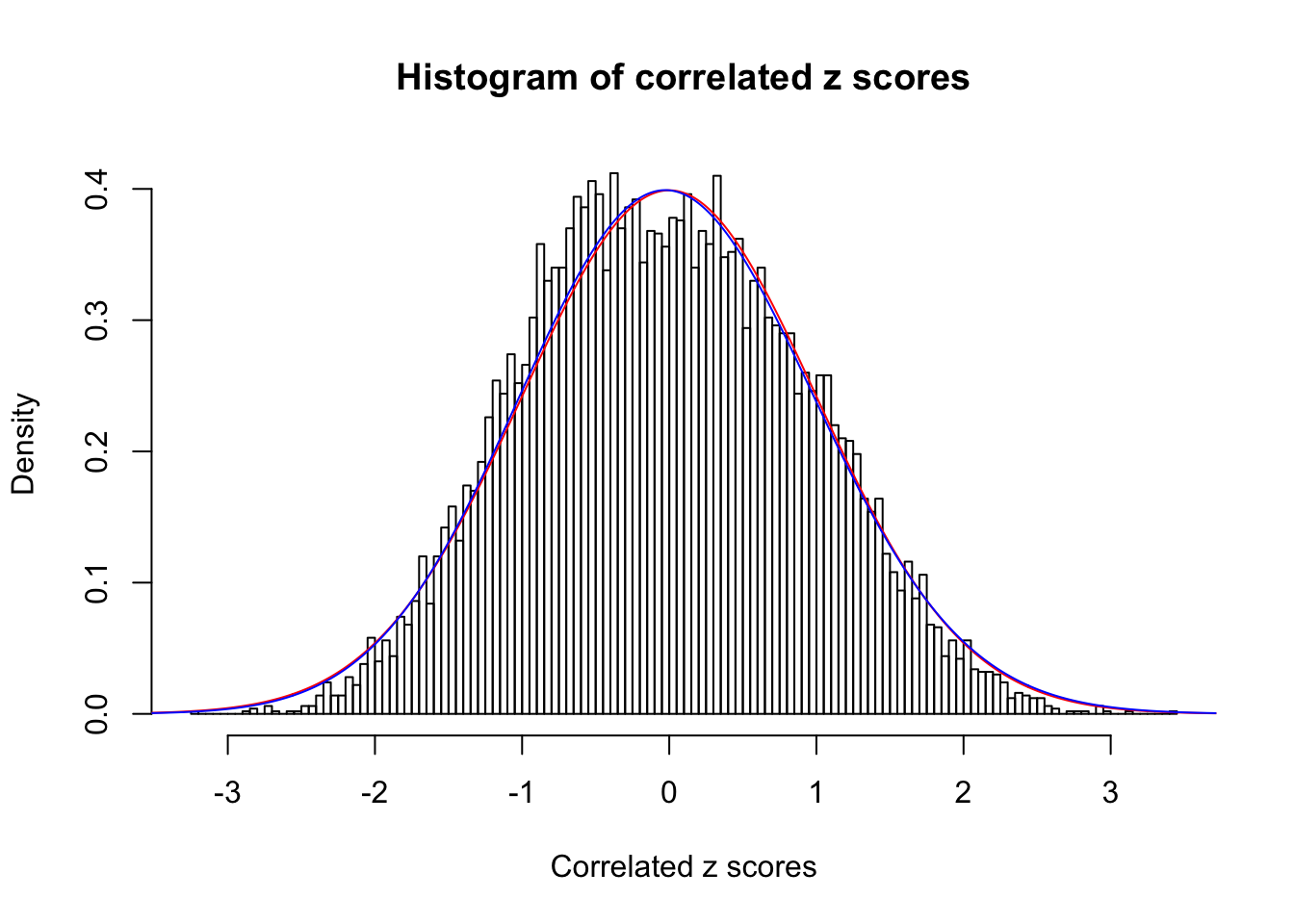
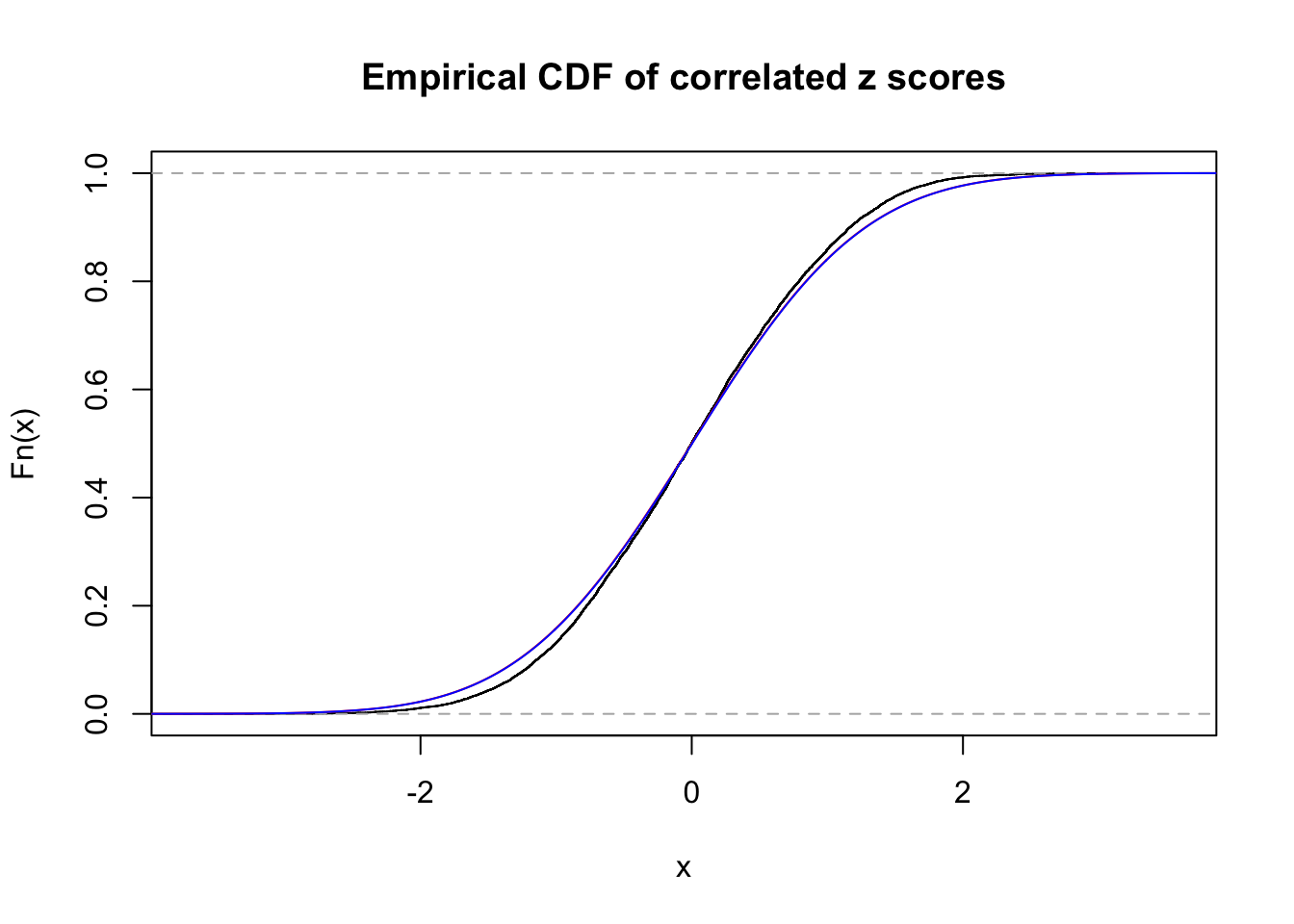
Data Set 5 :
Number of BH false discoveries: 0 ;
ASH's pihat0 = 1 ;
Normalized coefficients of Gaussian derivatives:
1 : 0.00299571447314808 ; 2 : 0 ; 3 : 0 ; 4 : 0 ;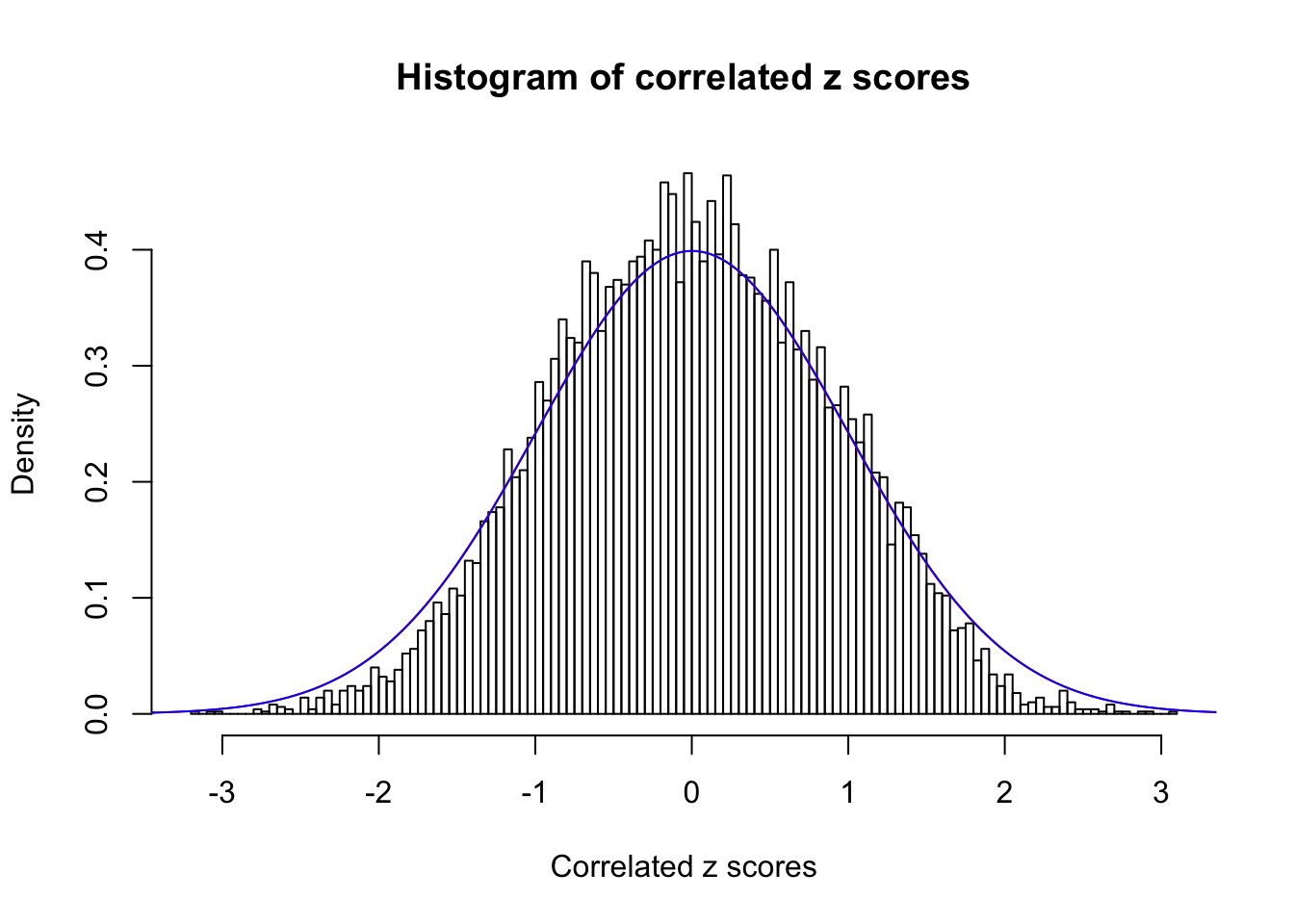
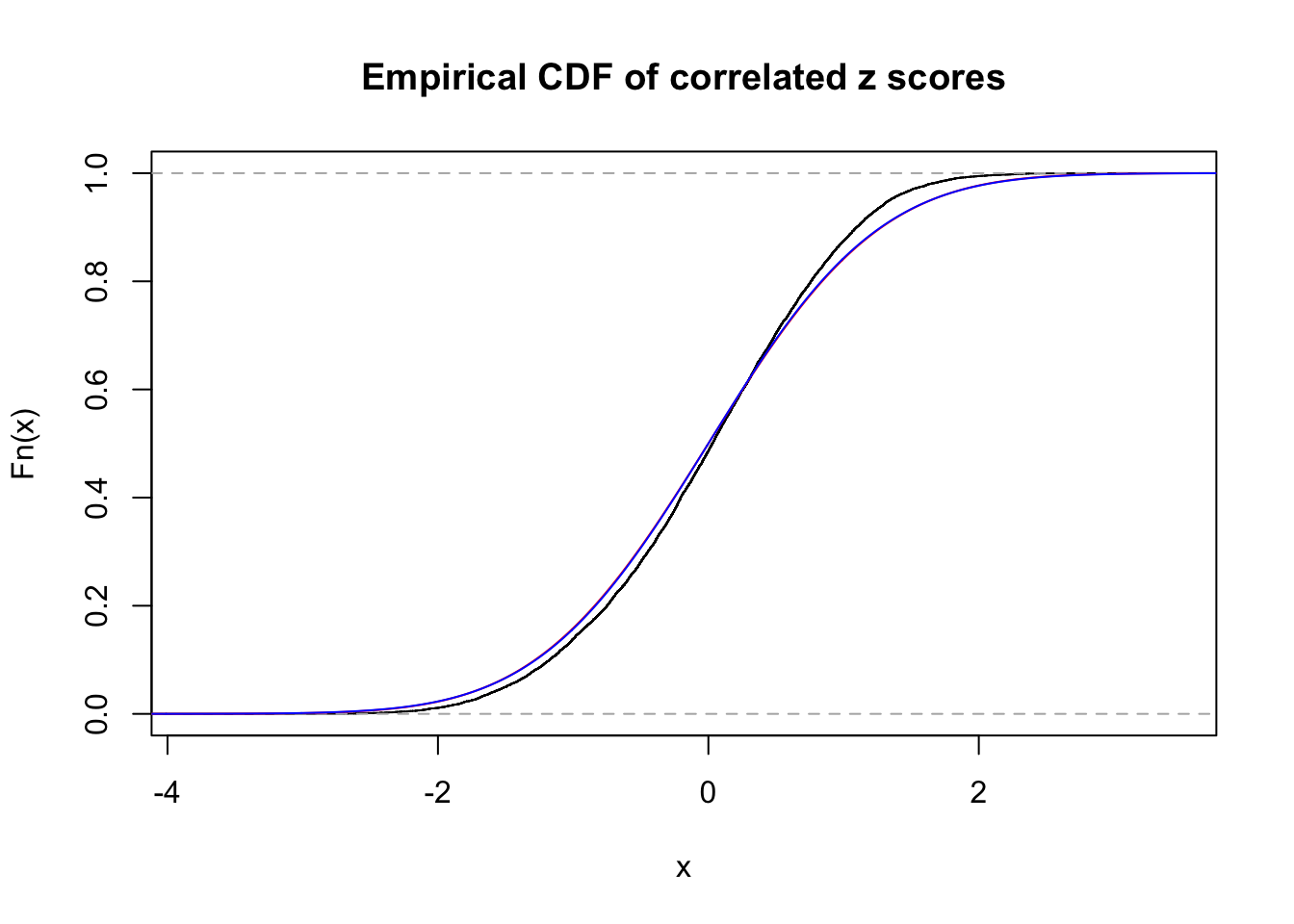
Data Set 7 :
Number of BH false discoveries: 0 ;
ASH's pihat0 = 1 ;
Normalized coefficients of Gaussian derivatives:
1 : 0 ; 2 : 0 ; 3 : 0 ; 4 : 0.0154789111560678 ;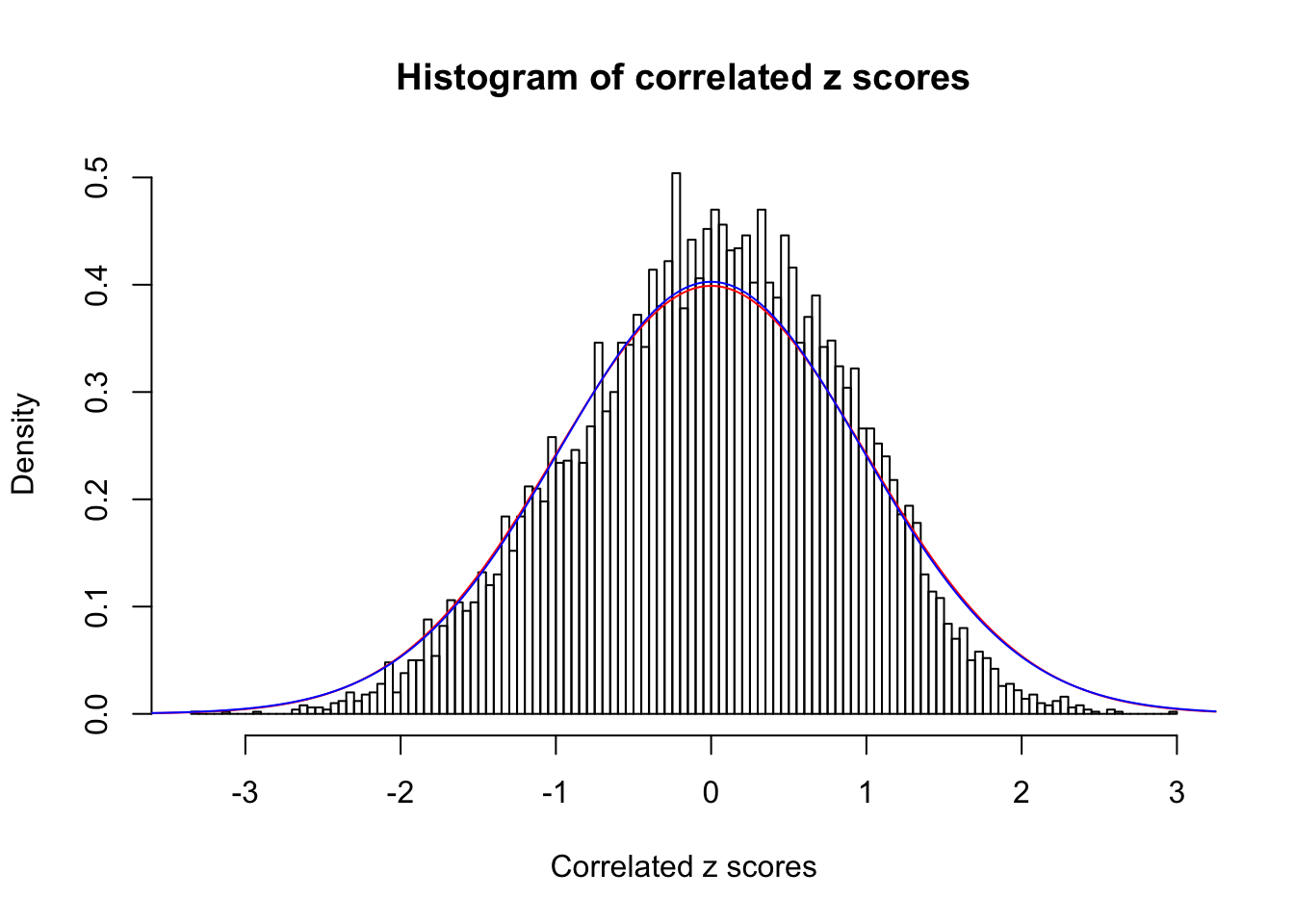
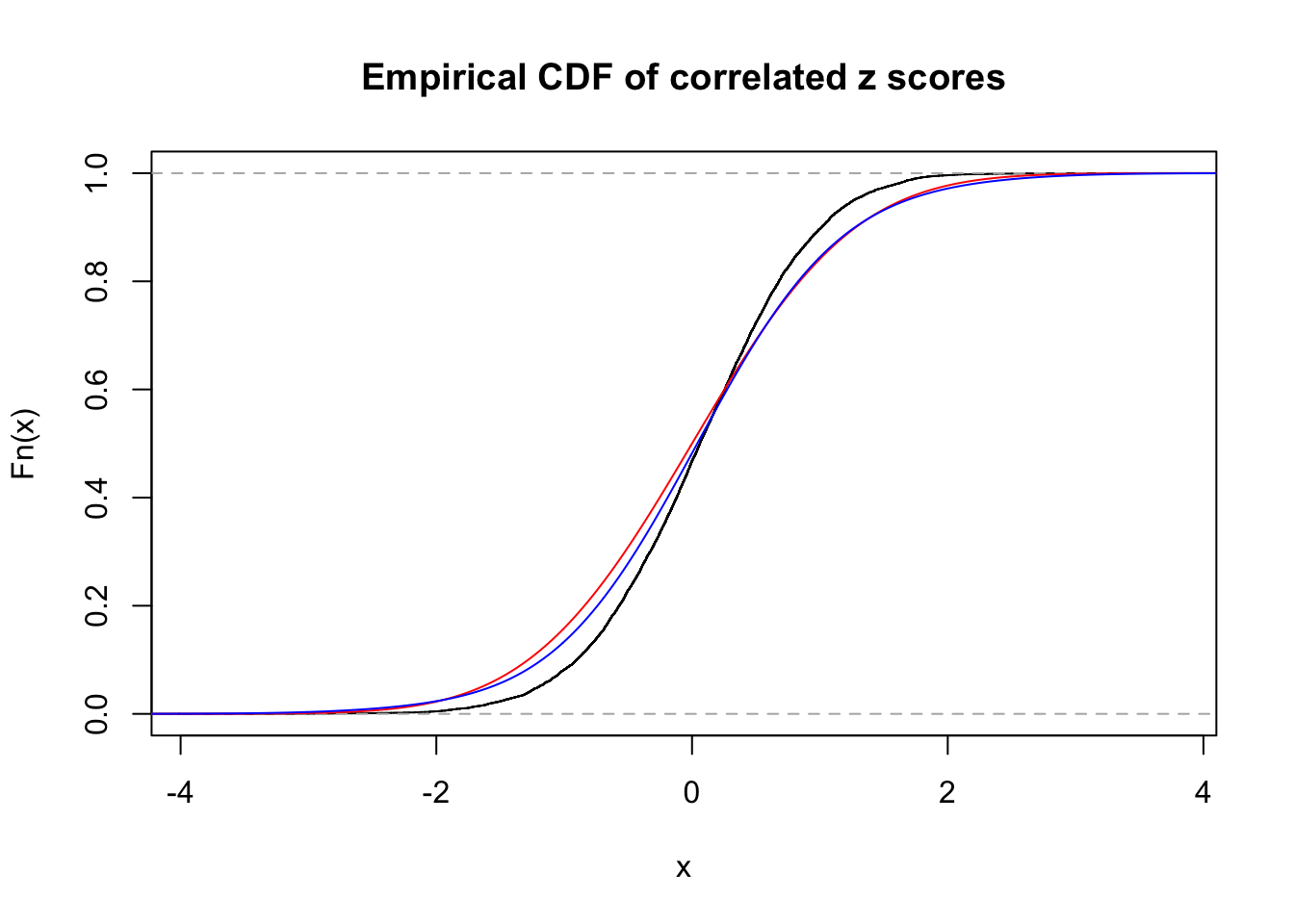
Data Set 8 :
Number of BH false discoveries: 0 ;
ASH's pihat0 = 1 ;
Normalized coefficients of Gaussian derivatives:
1 : 0.0448250444179864 ; 2 : 0 ; 3 : 0 ; 4 : 0.143559202208159 ;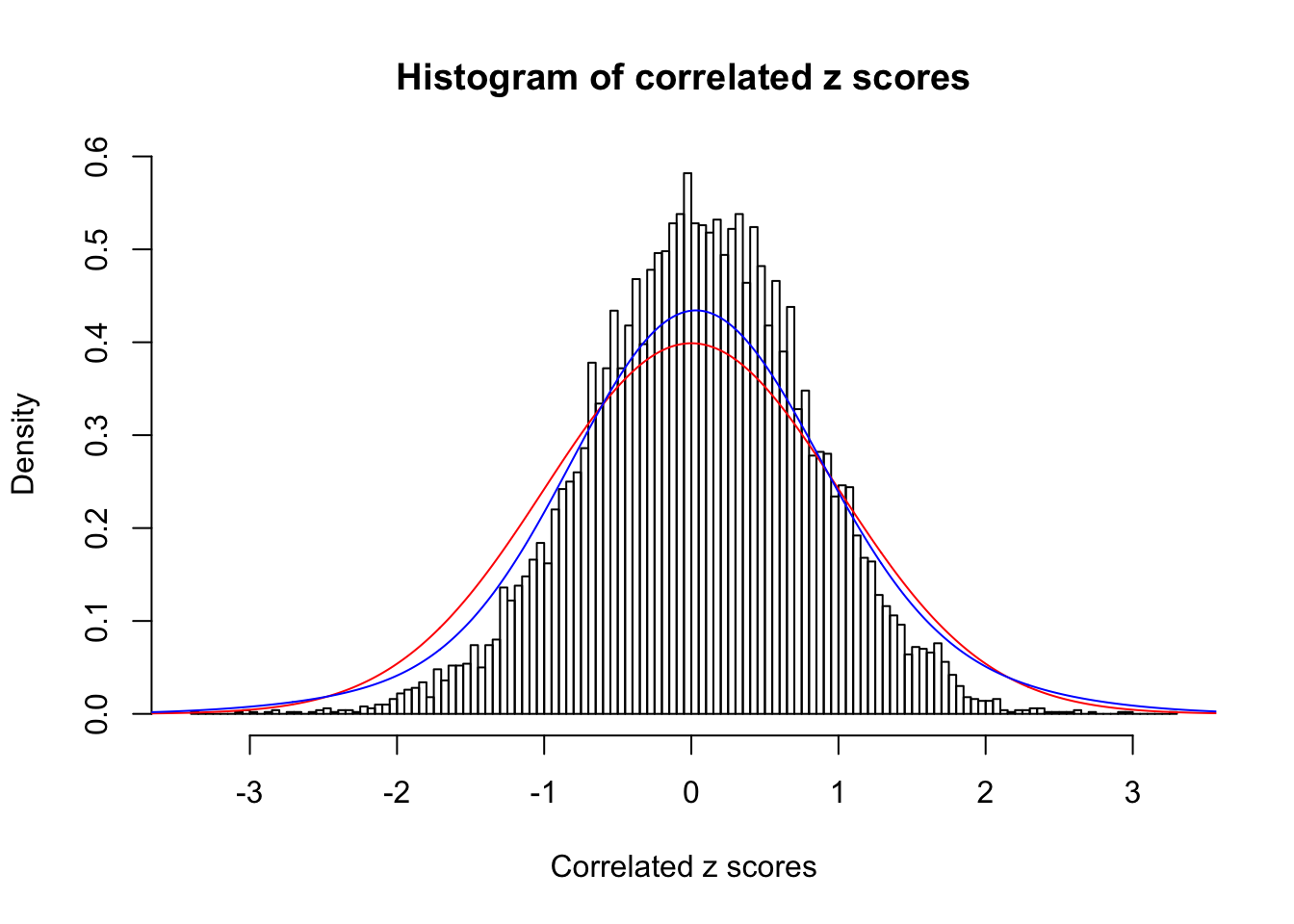
Data Set 9 :
Number of BH false discoveries: 0 ;
ASH's pihat0 = 1 ;
Normalized coefficients of Gaussian derivatives:
1 : 0 ; 2 : 0.00733790864173648 ; 3 : 0.0163403533804334 ; 4 : 0 ;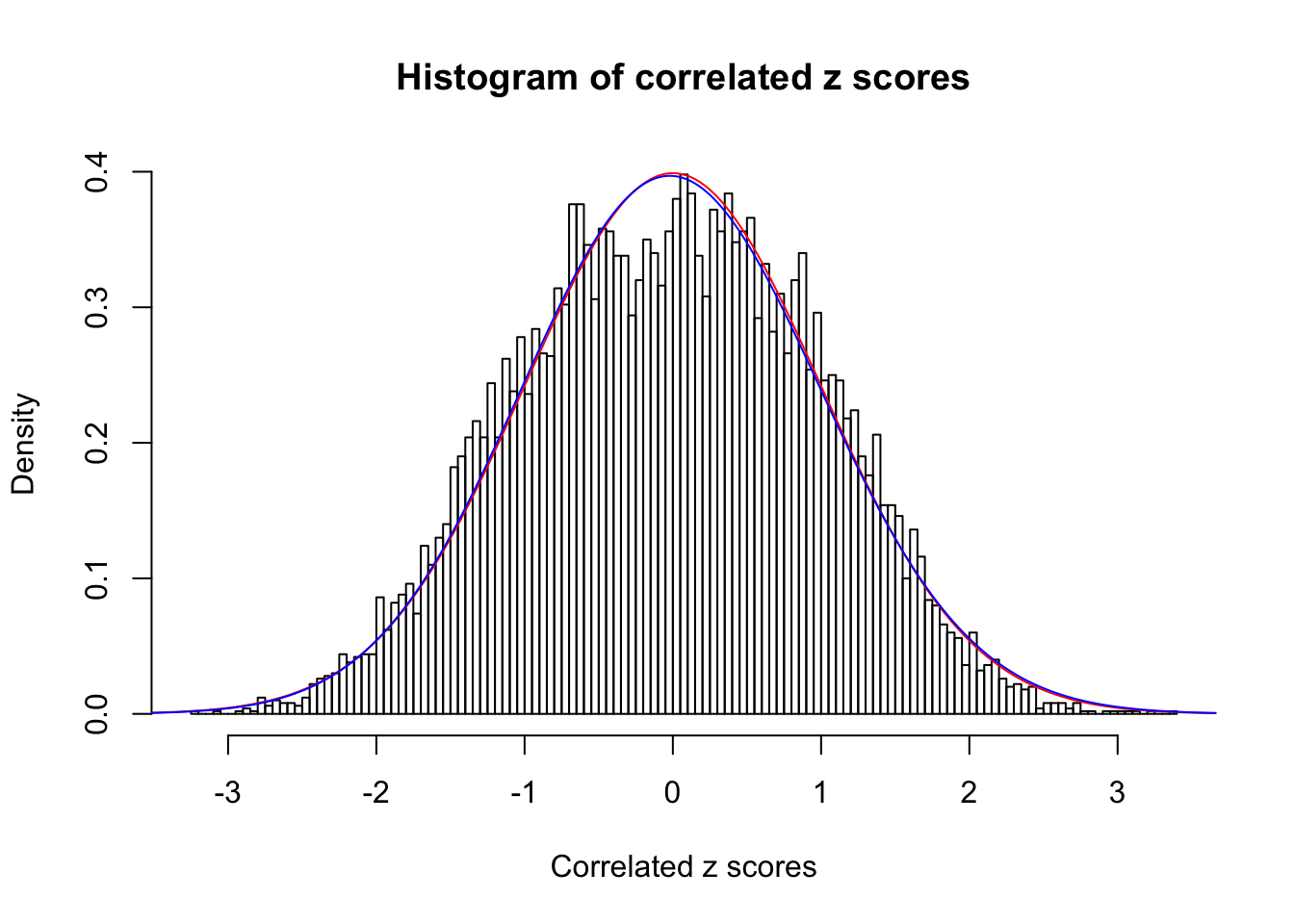
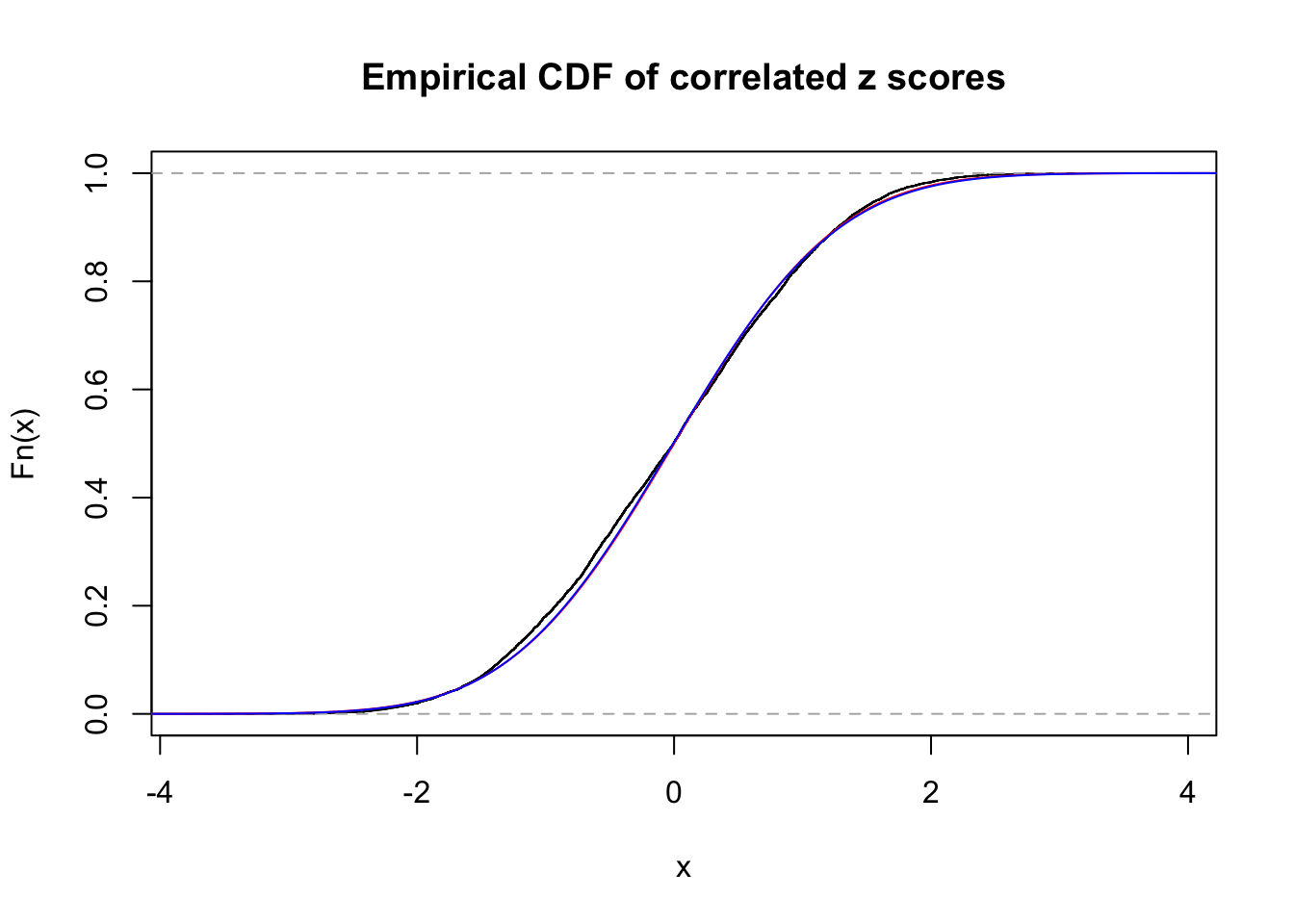
Data Set 12 :
Number of BH false discoveries: 0 ;
ASH's pihat0 = 1 ;
Normalized coefficients of Gaussian derivatives:
1 : 0 ; 2 : 0 ; 3 : 0 ; 4 : 0.0221346391841642 ;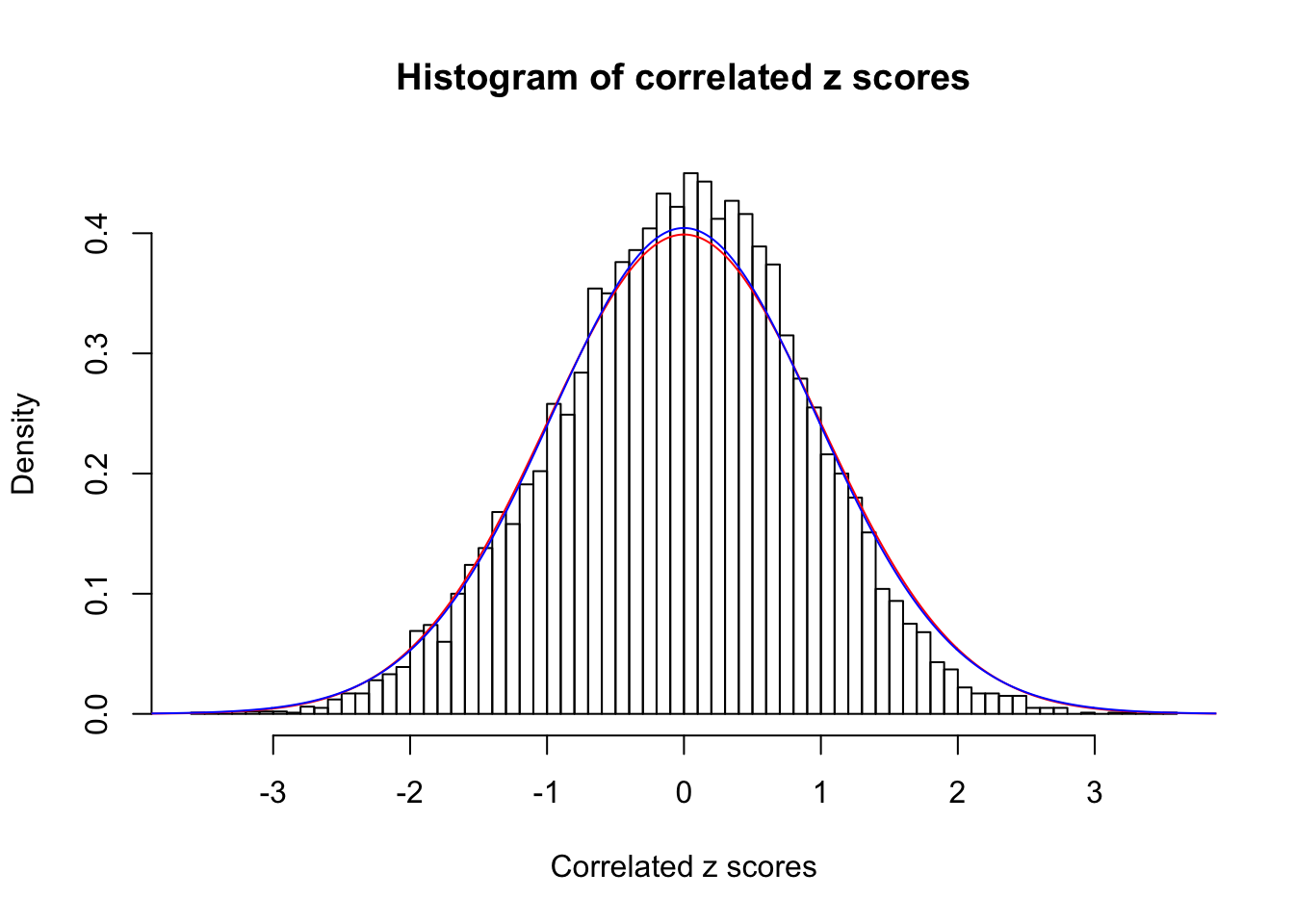
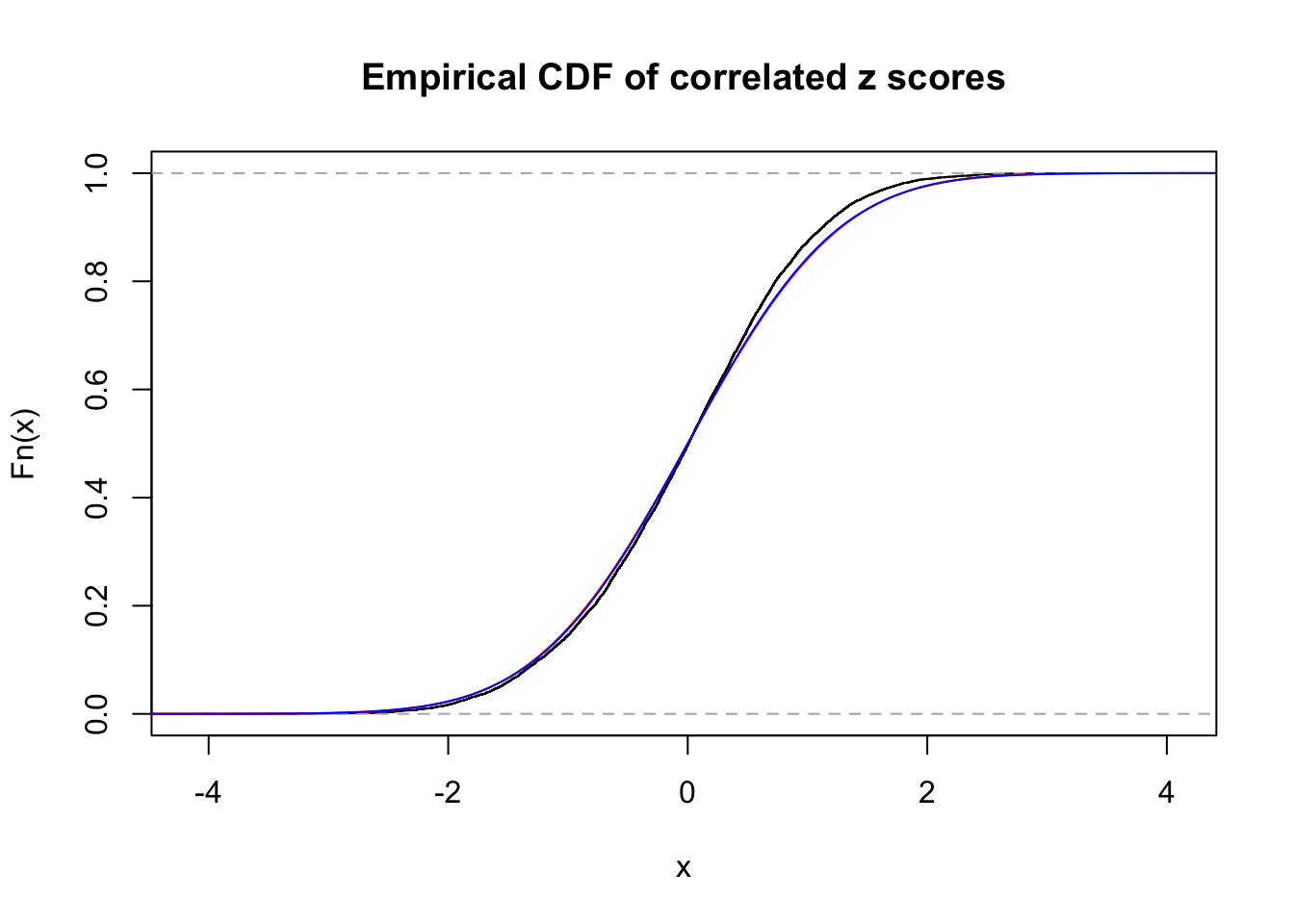
Data Set 13 :
Number of BH false discoveries: 0 ;
ASH's pihat0 = 1 ;
Normalized coefficients of Gaussian derivatives:
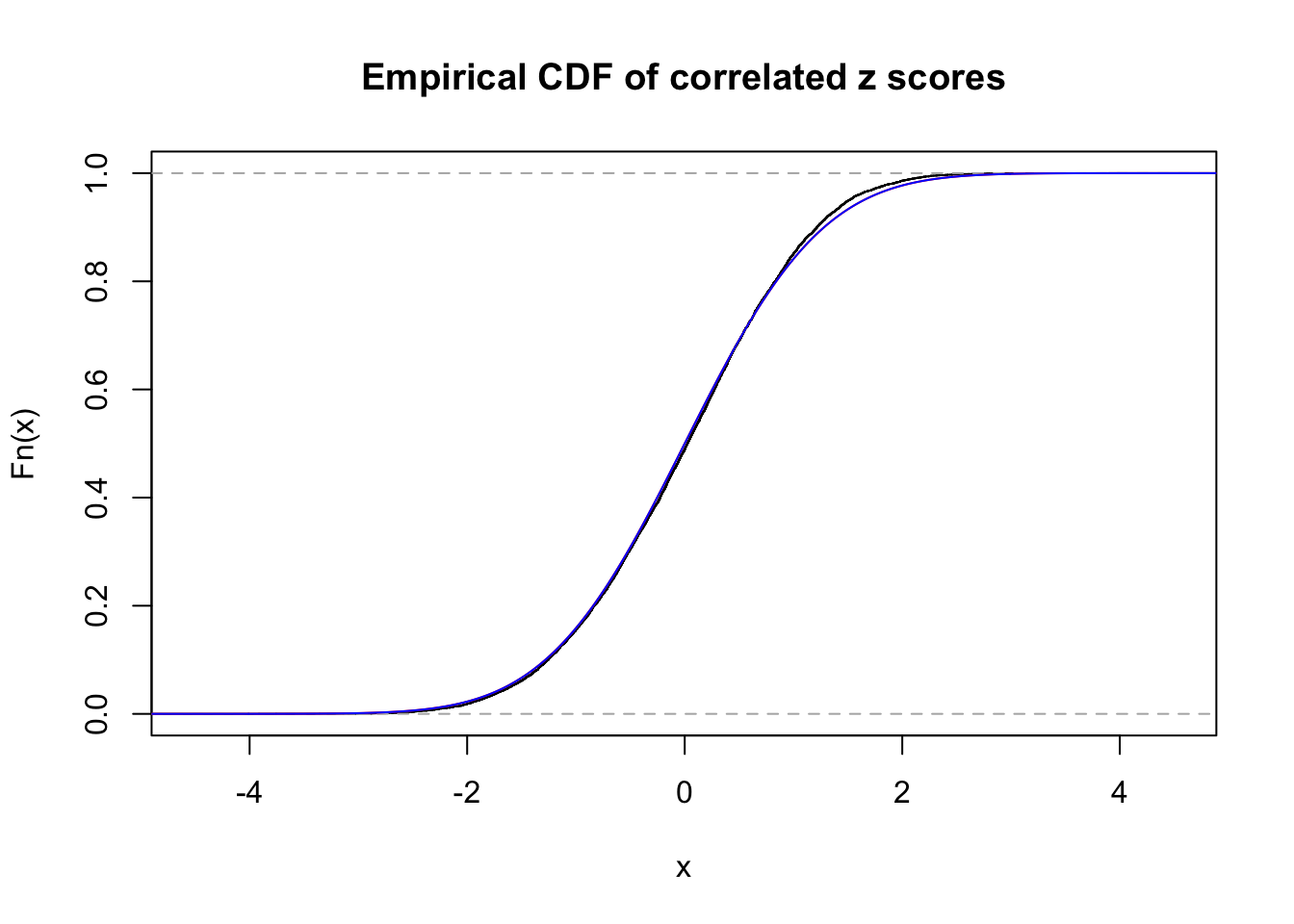
1 : 0 ; 2 : 0 ; 3 : 0 ; 4 : 0 ;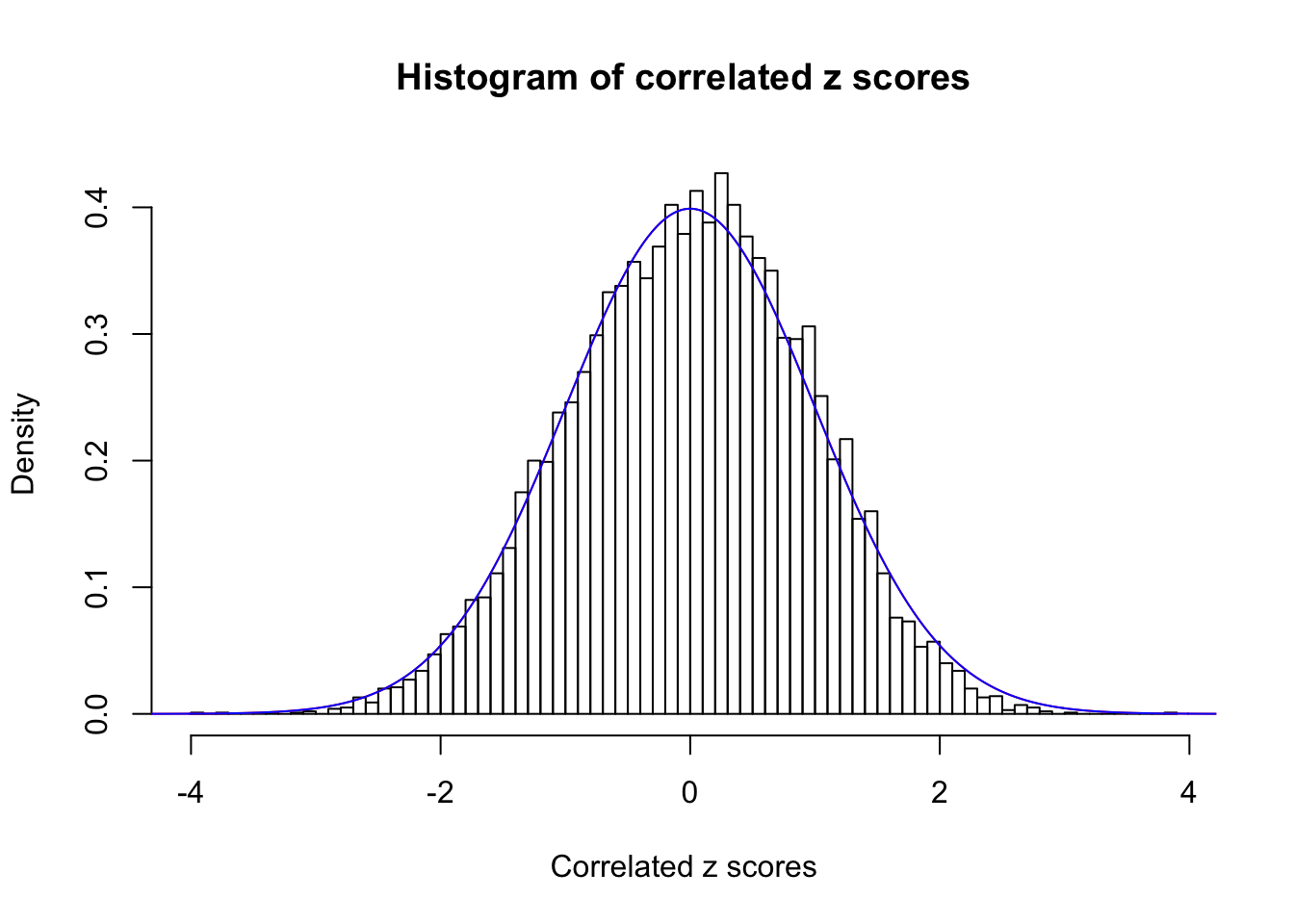
Data Set 14 :
Number of BH false discoveries: 0 ;
ASH's pihat0 = 1 ;
Normalized coefficients of Gaussian derivatives:
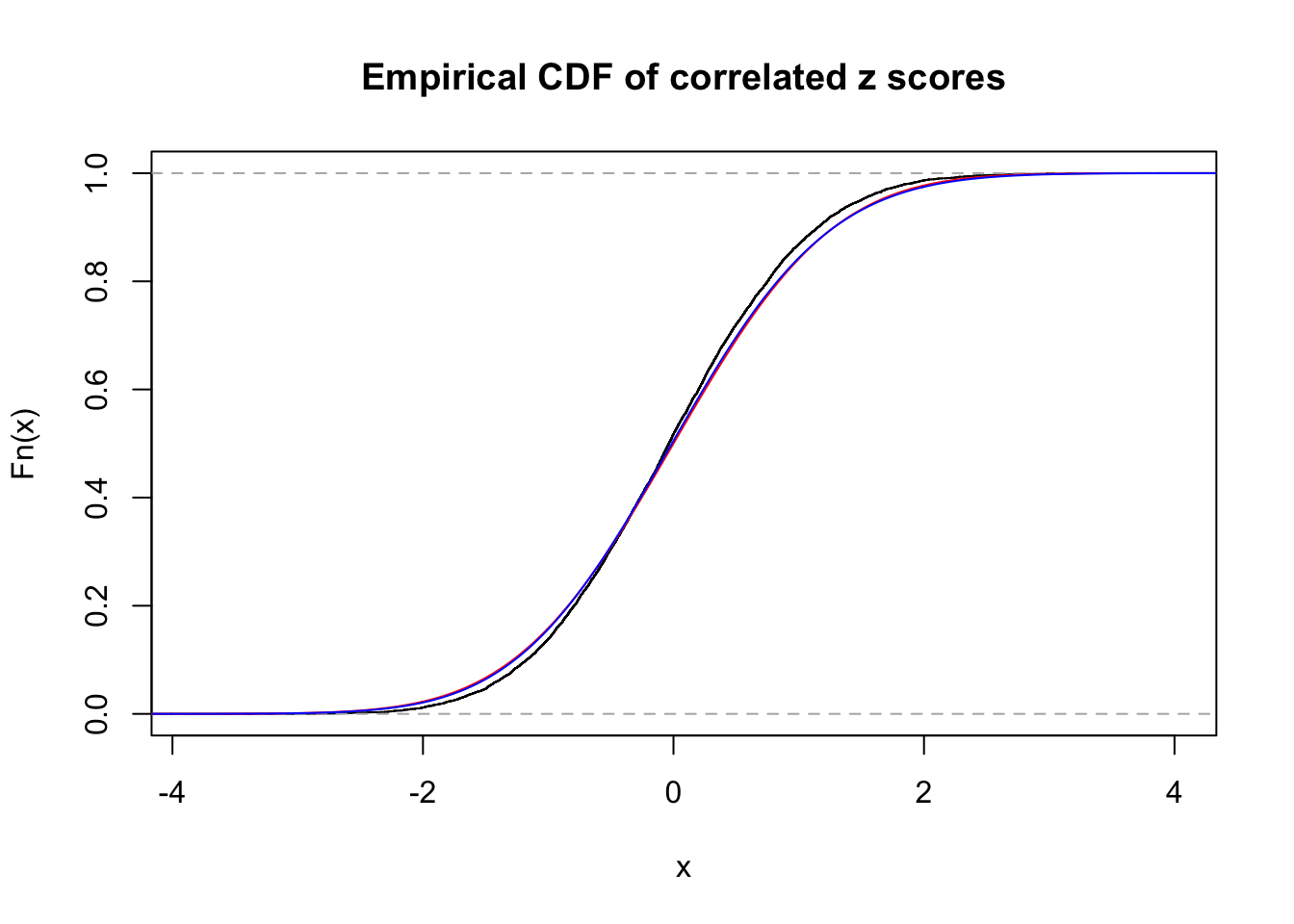
1 : 0 ; 2 : 0 ; 3 : 0.0319220622489123 ; 4 : 0.0192425975845108 ;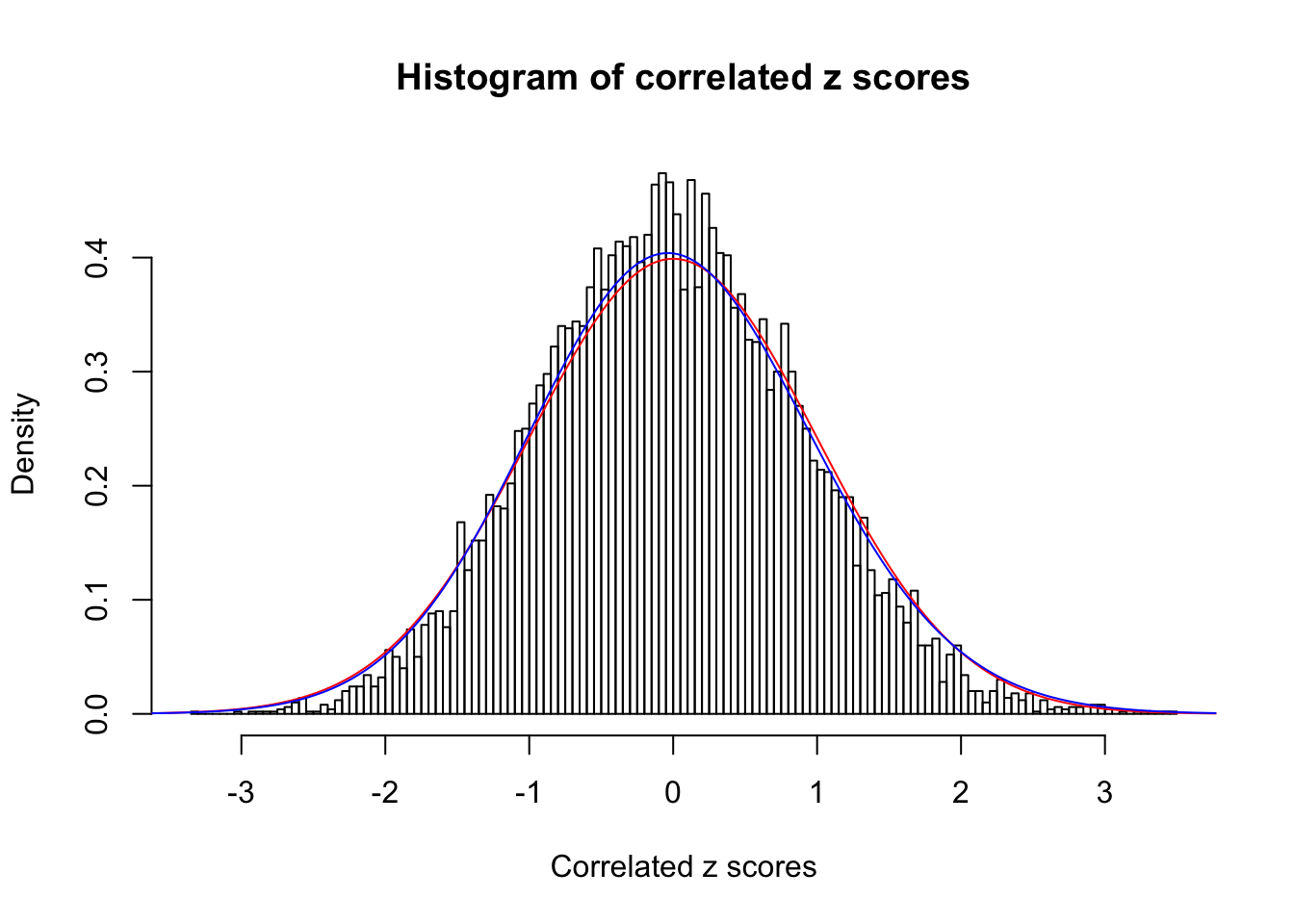
Data Set 16 :
Number of BH false discoveries: 0 ;
ASH's pihat0 = 1 ;
Normalized coefficients of Gaussian derivatives:
1 : 0.0132467627058809 ; 2 : 0 ; 3 : 0.0050633052630629 ; 4 : 0.0460280223671055 ;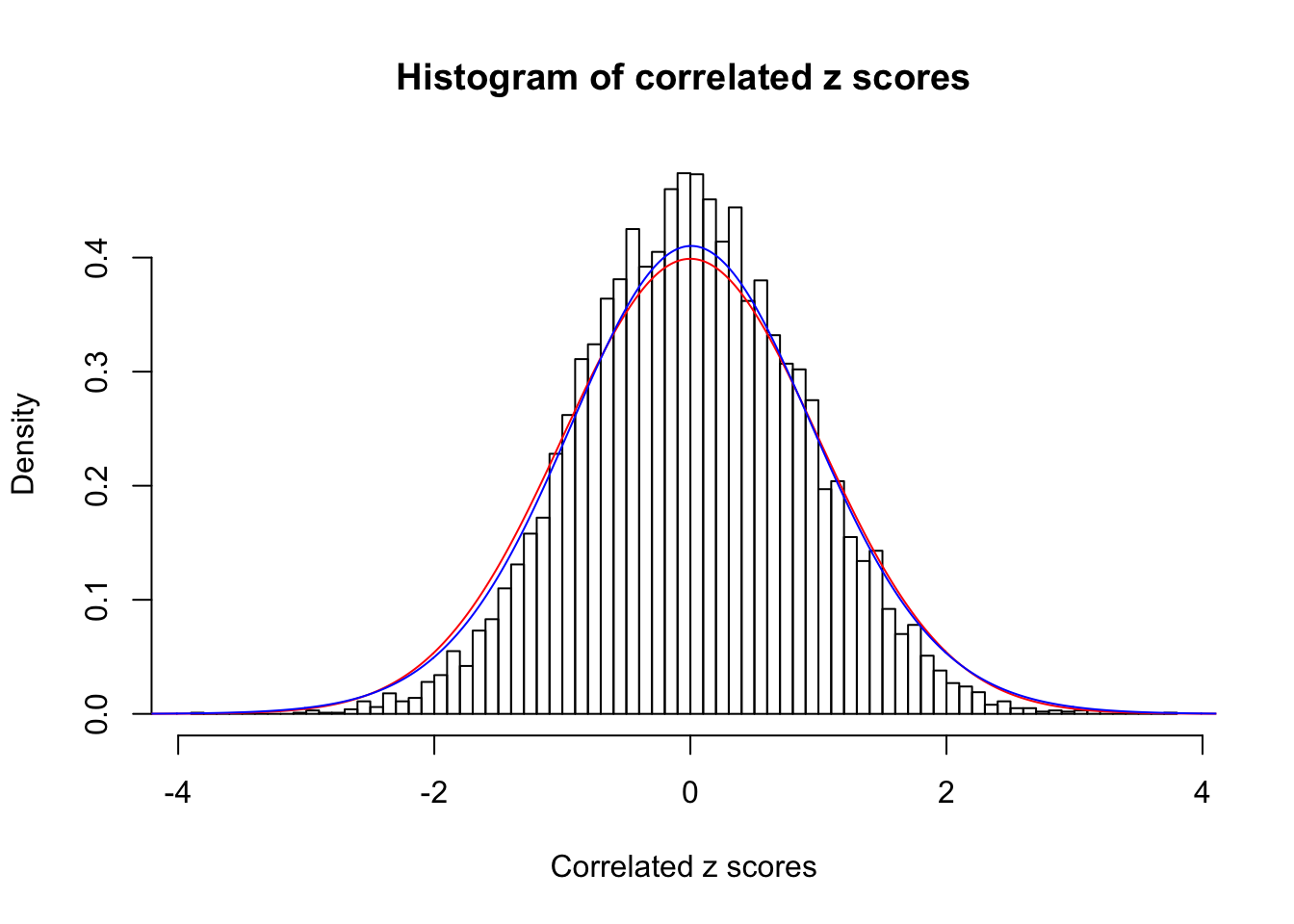
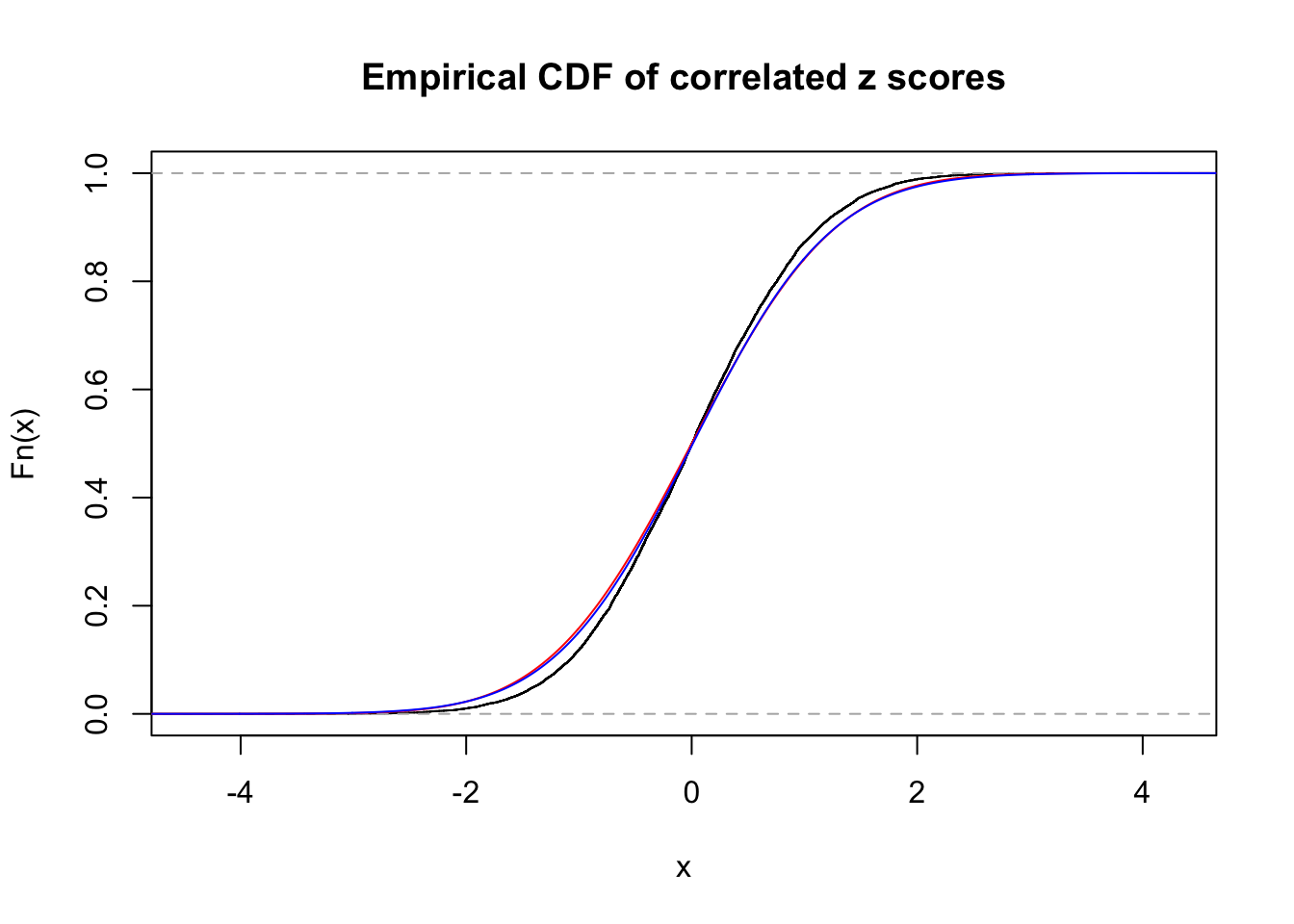
Data Set 17 :
Number of BH false discoveries: 0 ;
ASH's pihat0 = 1 ;
Normalized coefficients of Gaussian derivatives:
1 : 0.019759781277875 ; 2 : 0 ; 3 : 0 ; 4 : 0.0663856267063882 ;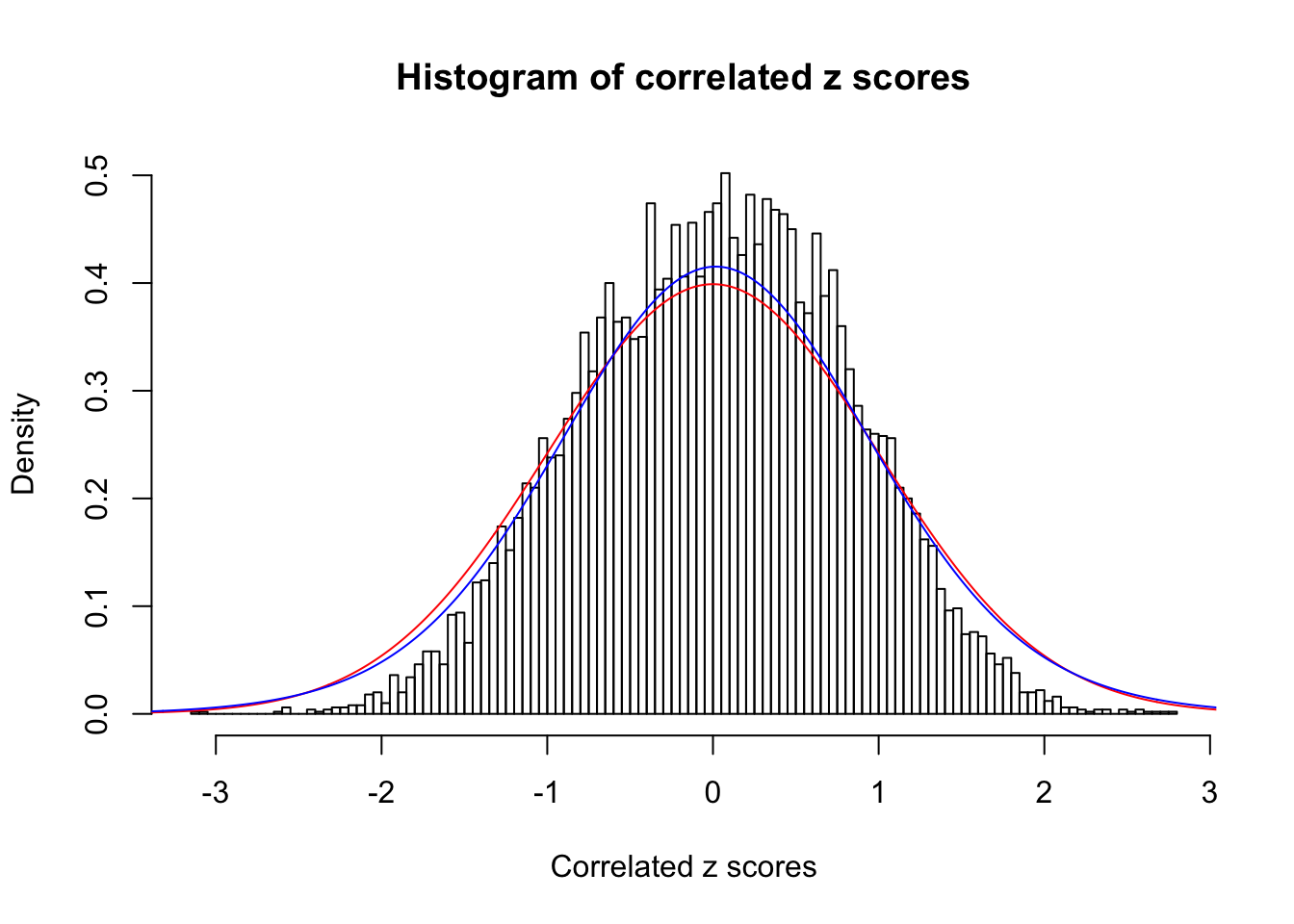
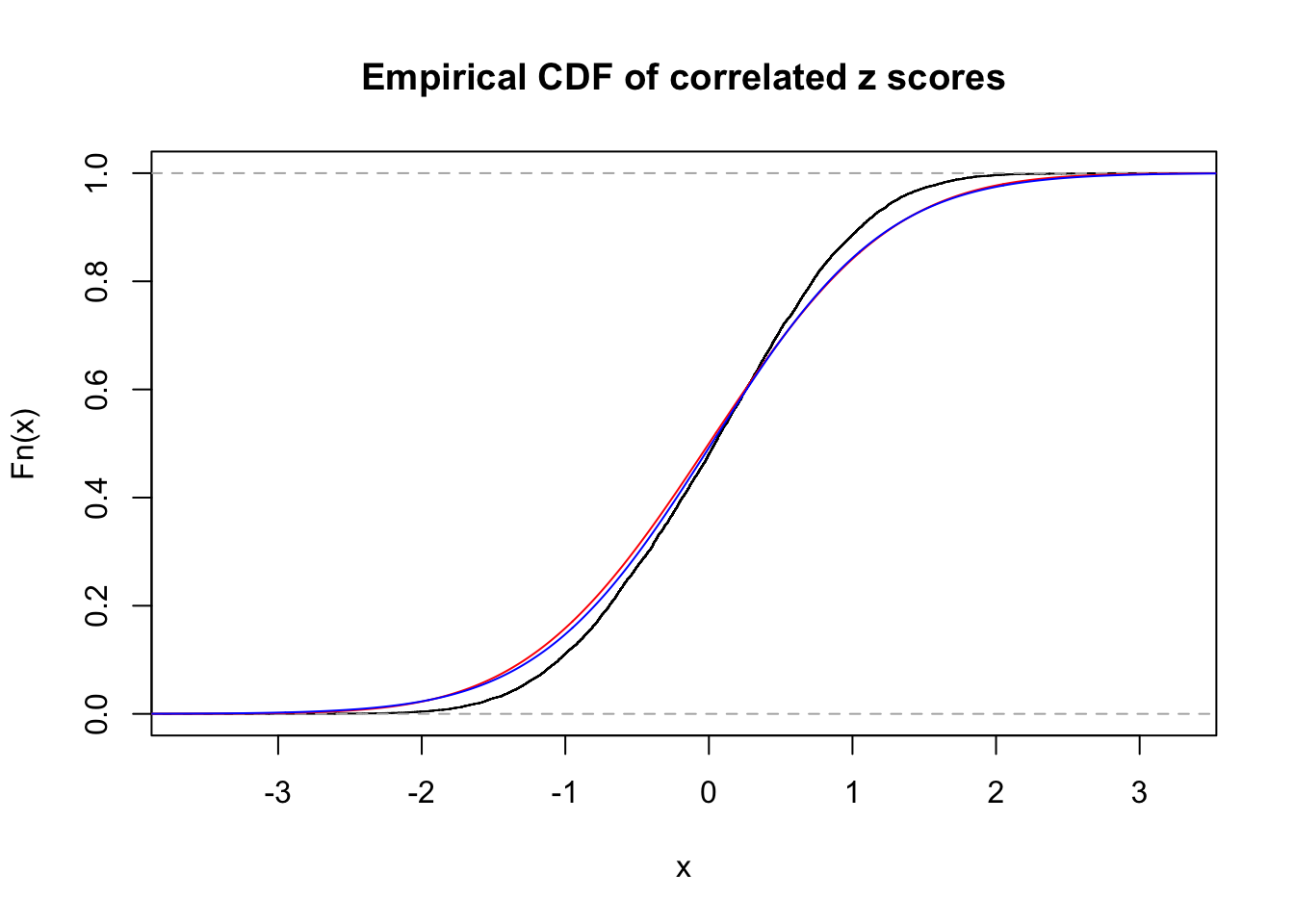
Data Set 18 :
Number of BH false discoveries: 0 ;
ASH's pihat0 = 1 ;
Normalized coefficients of Gaussian derivatives:
1 : 0 ; 2 : 0 ; 3 : 0.0334151016576349 ; 4 : 0.00459370714209619 ;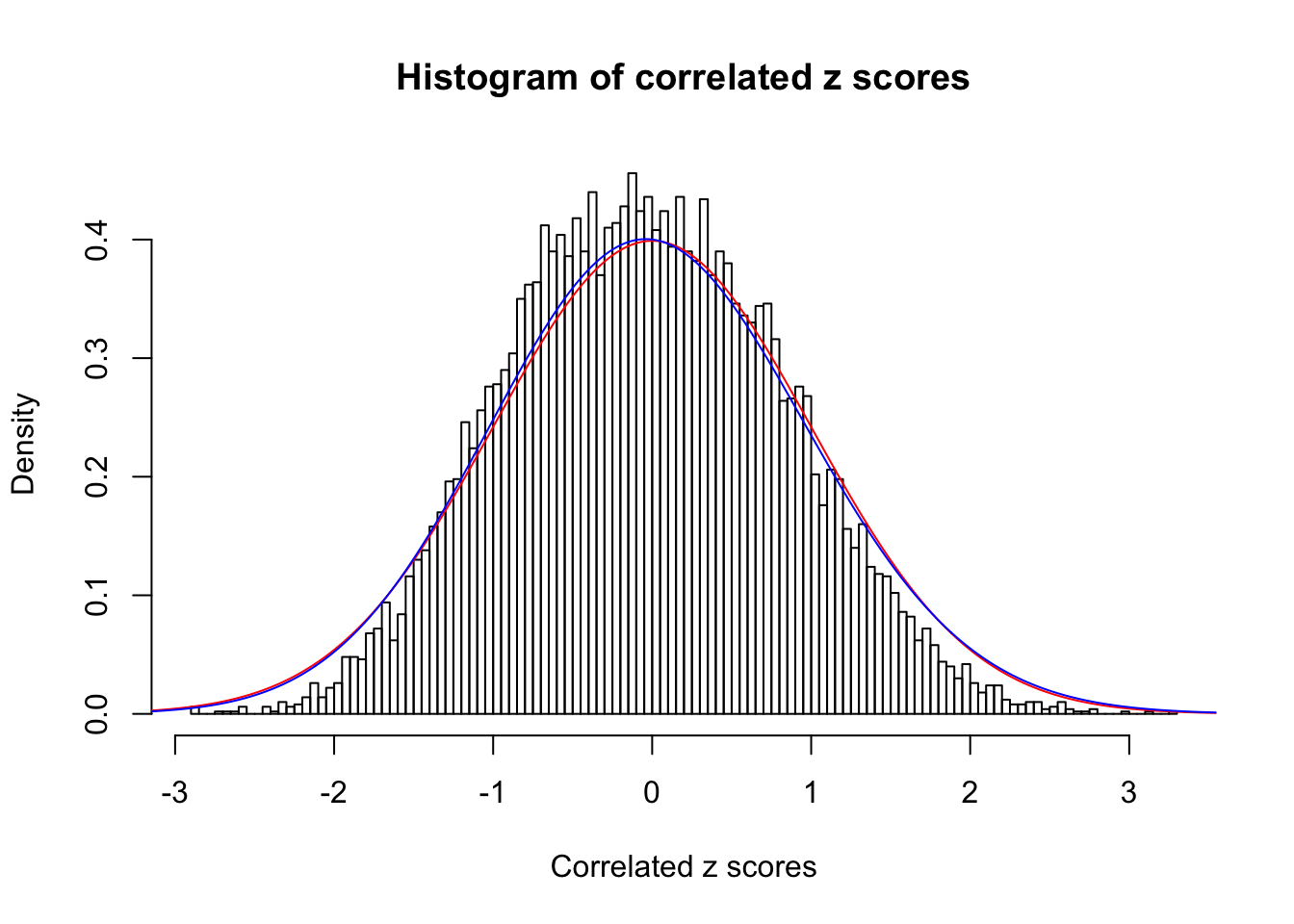
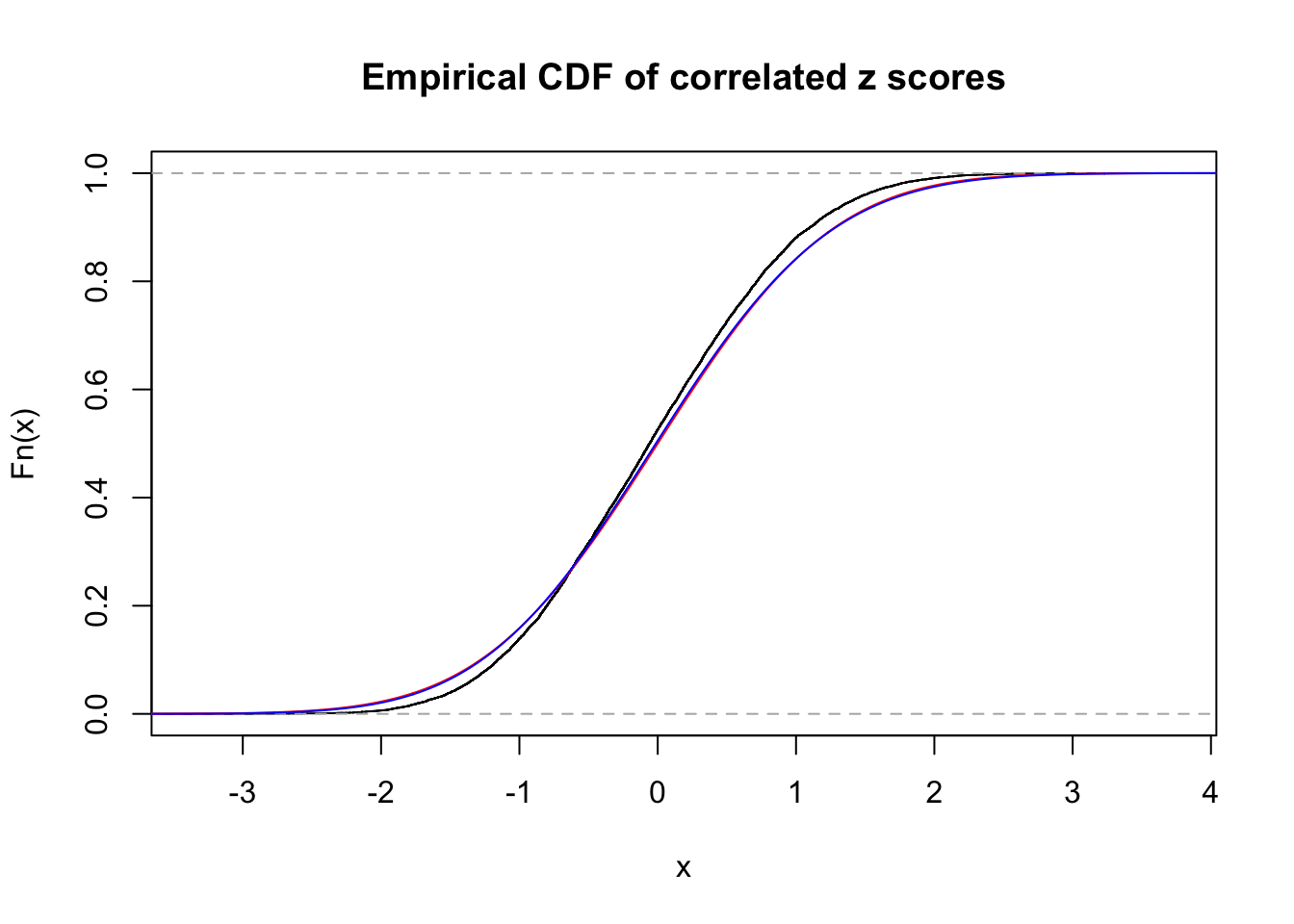
Data Set 25 :
Number of BH false discoveries: 0 ;
ASH's pihat0 = 1 ;
Normalized coefficients of Gaussian derivatives:
1 : 0 ; 2 : 0 ; 3 : 0.0110557207434819 ; 4 : 0.165219572806289 ;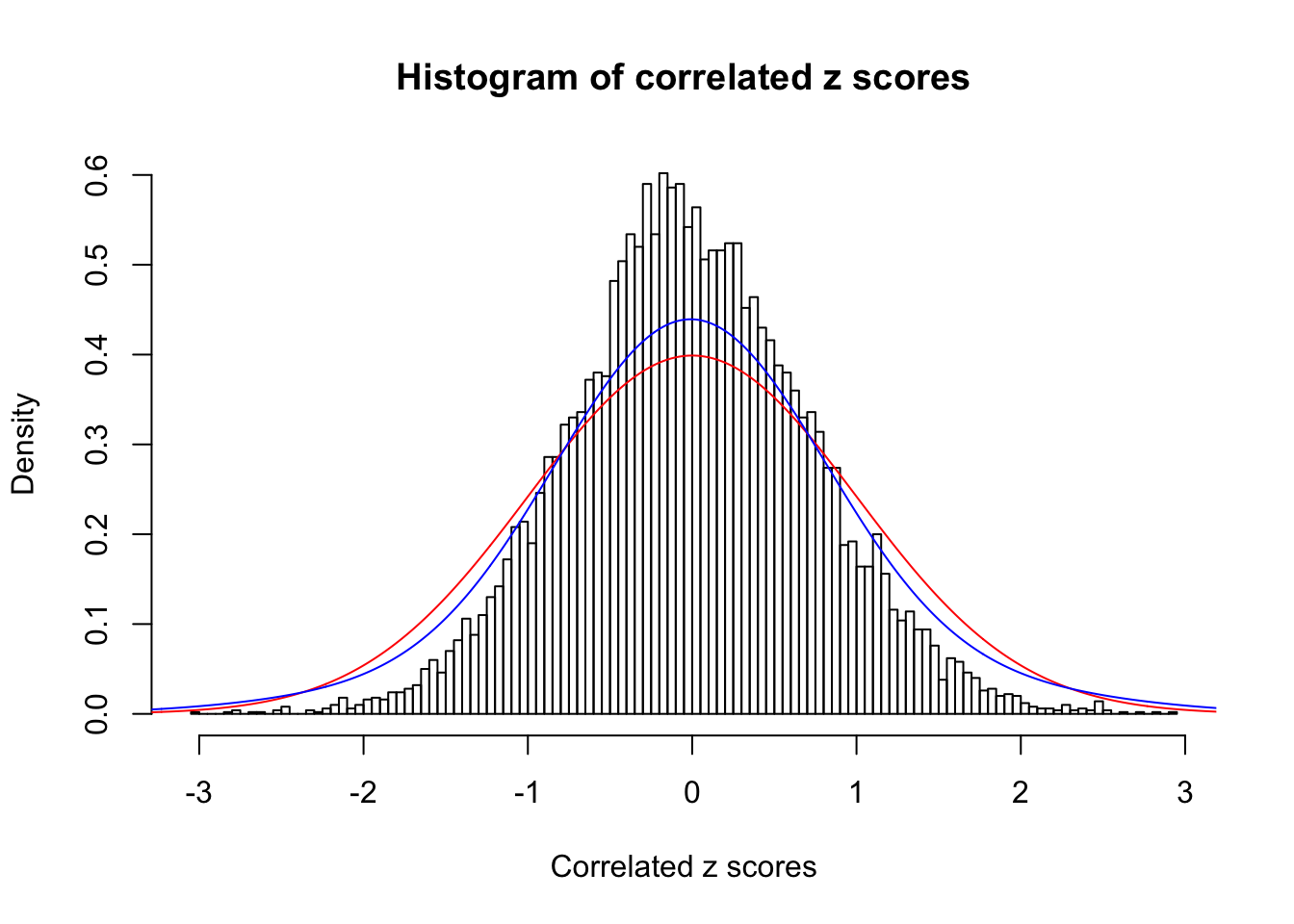
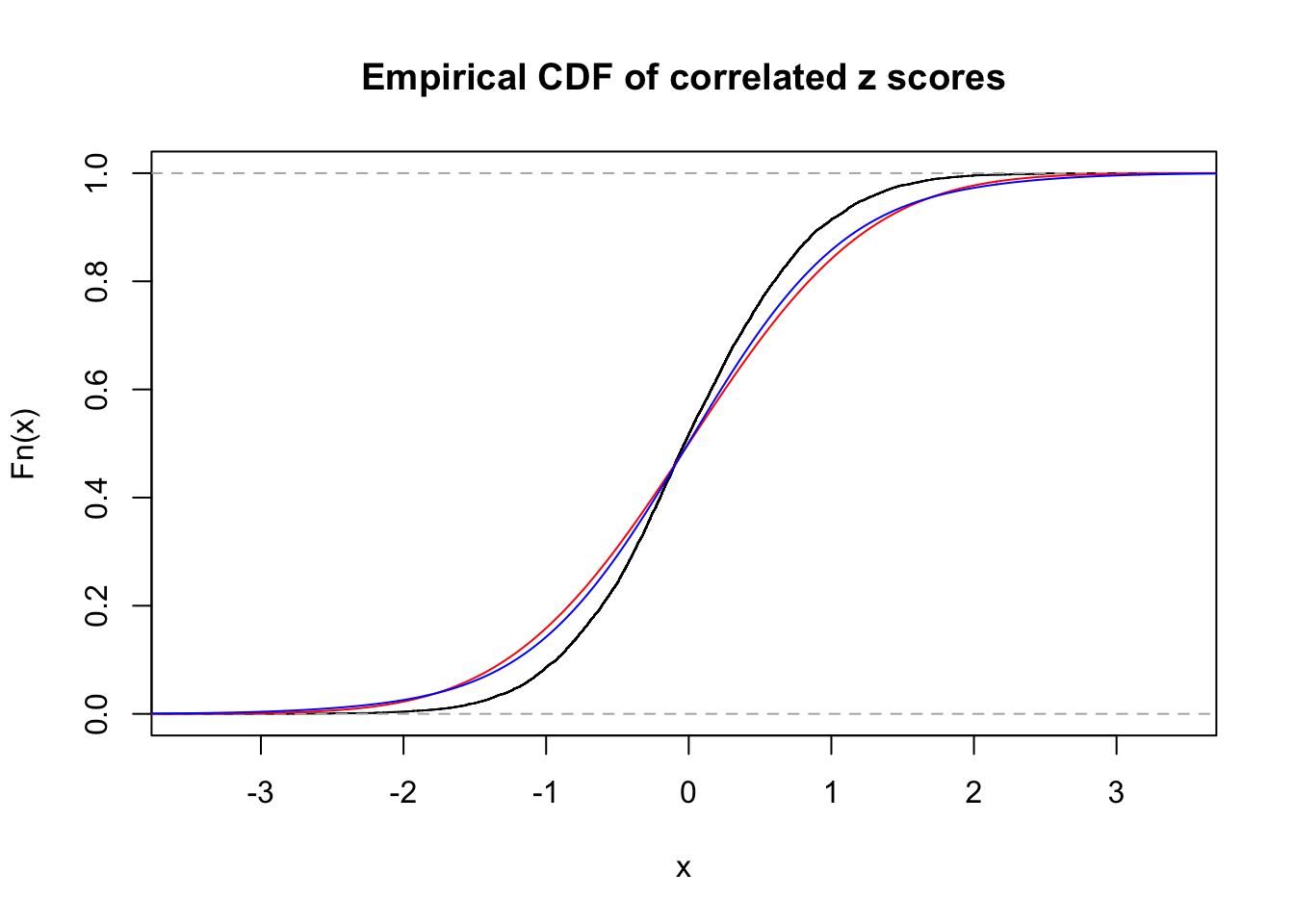
Data Set 28 :
Number of BH false discoveries: 0 ;
ASH's pihat0 = 1 ;
Normalized coefficients of Gaussian derivatives:
1 : 0.0200505087249252 ; 2 : 0 ; 3 : 0 ; 4 : 0 ;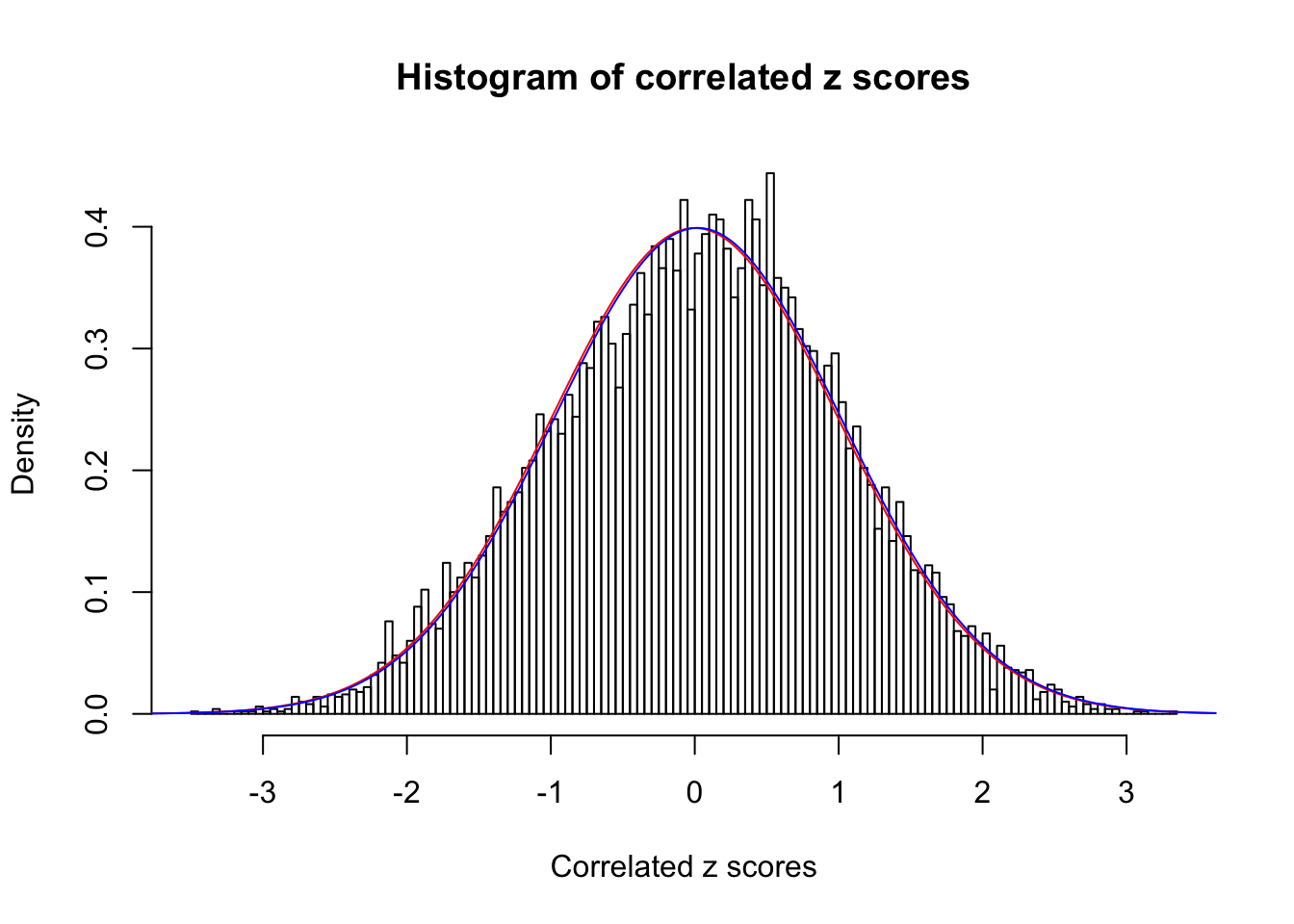
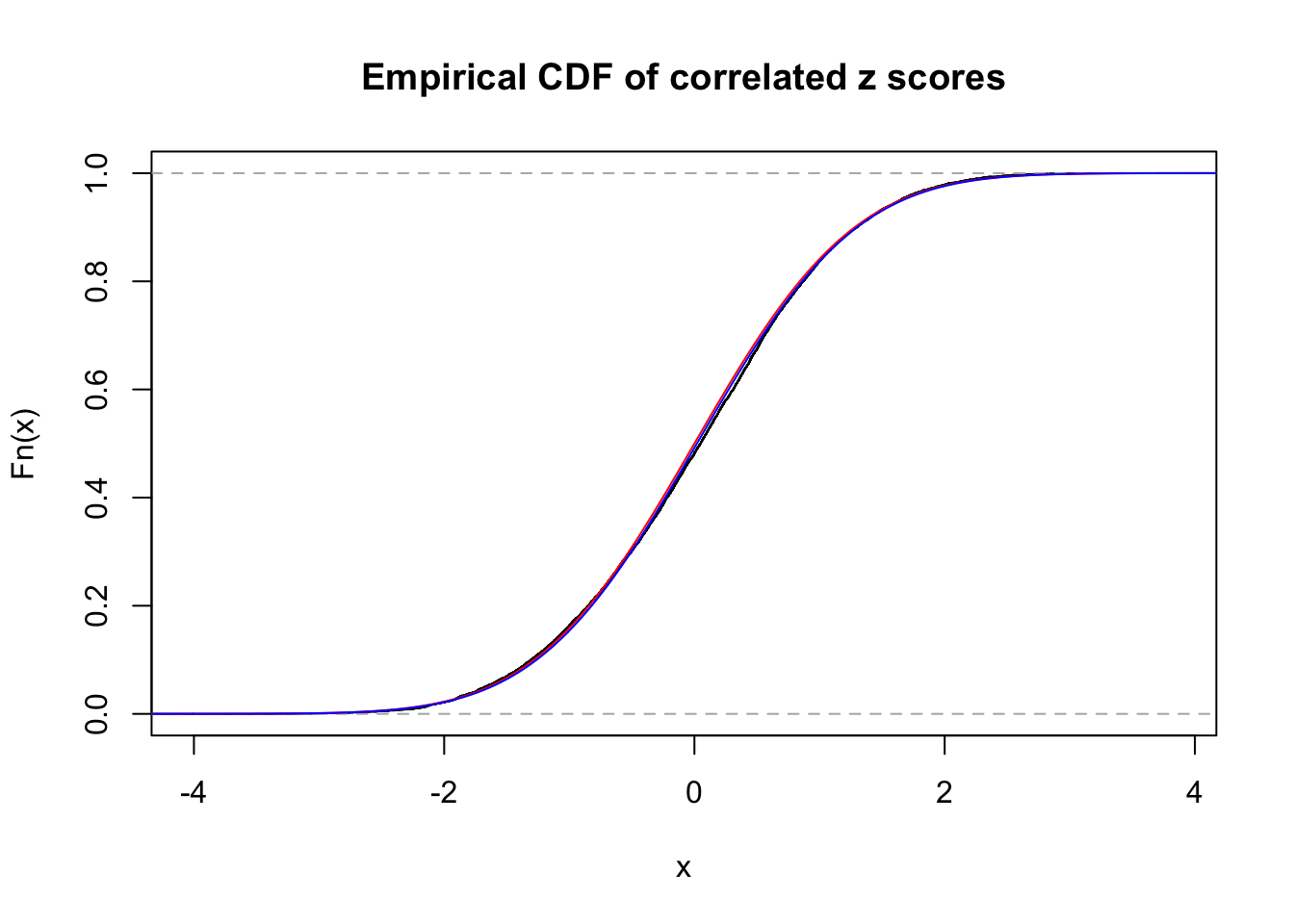
Data Set 30 :
Number of BH false discoveries: 0 ;
ASH's pihat0 = 1 ;
Normalized coefficients of Gaussian derivatives:
1 : 0 ; 2 : 0 ; 3 : 0.0118483507943389 ; 4 : 0 ;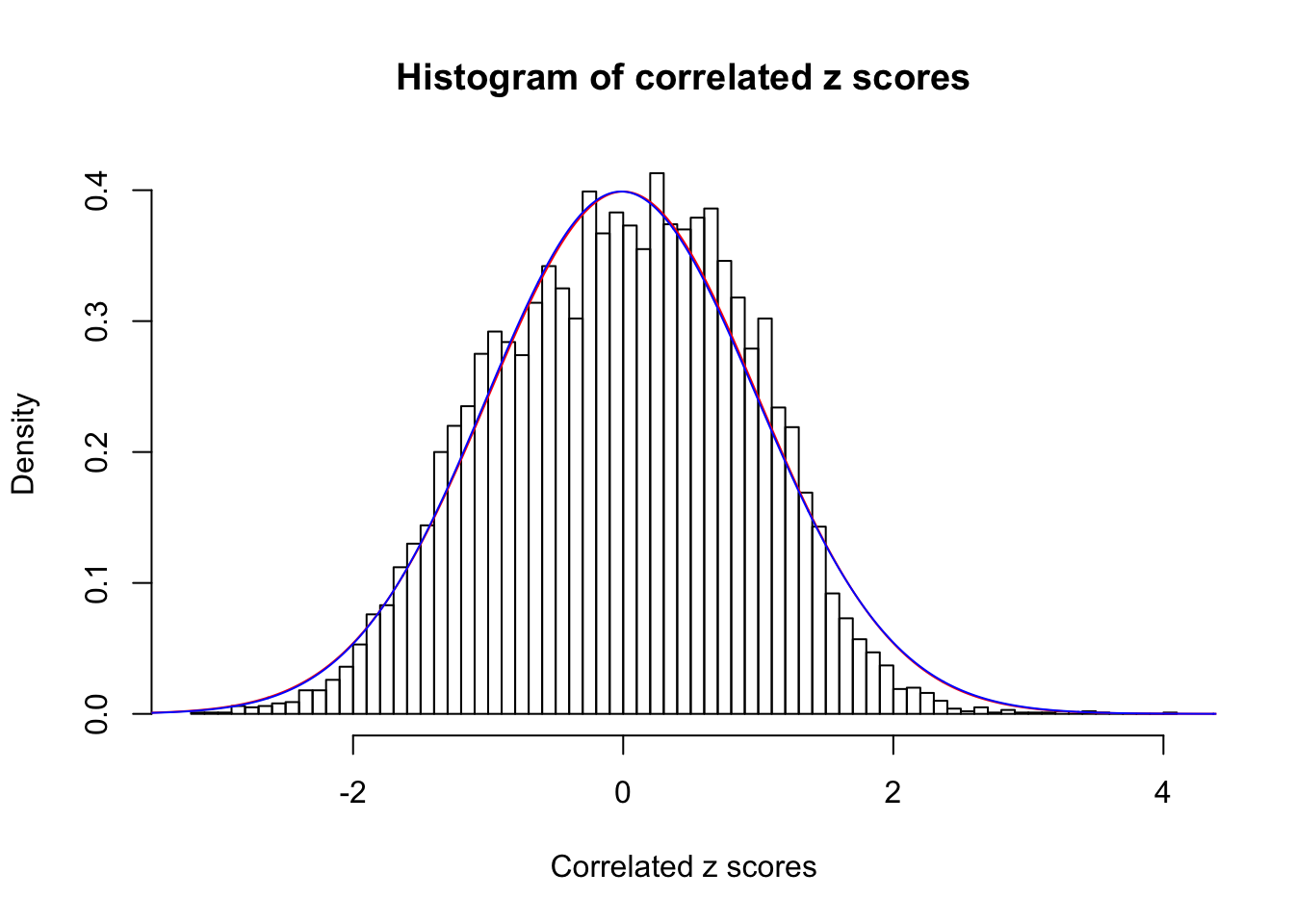
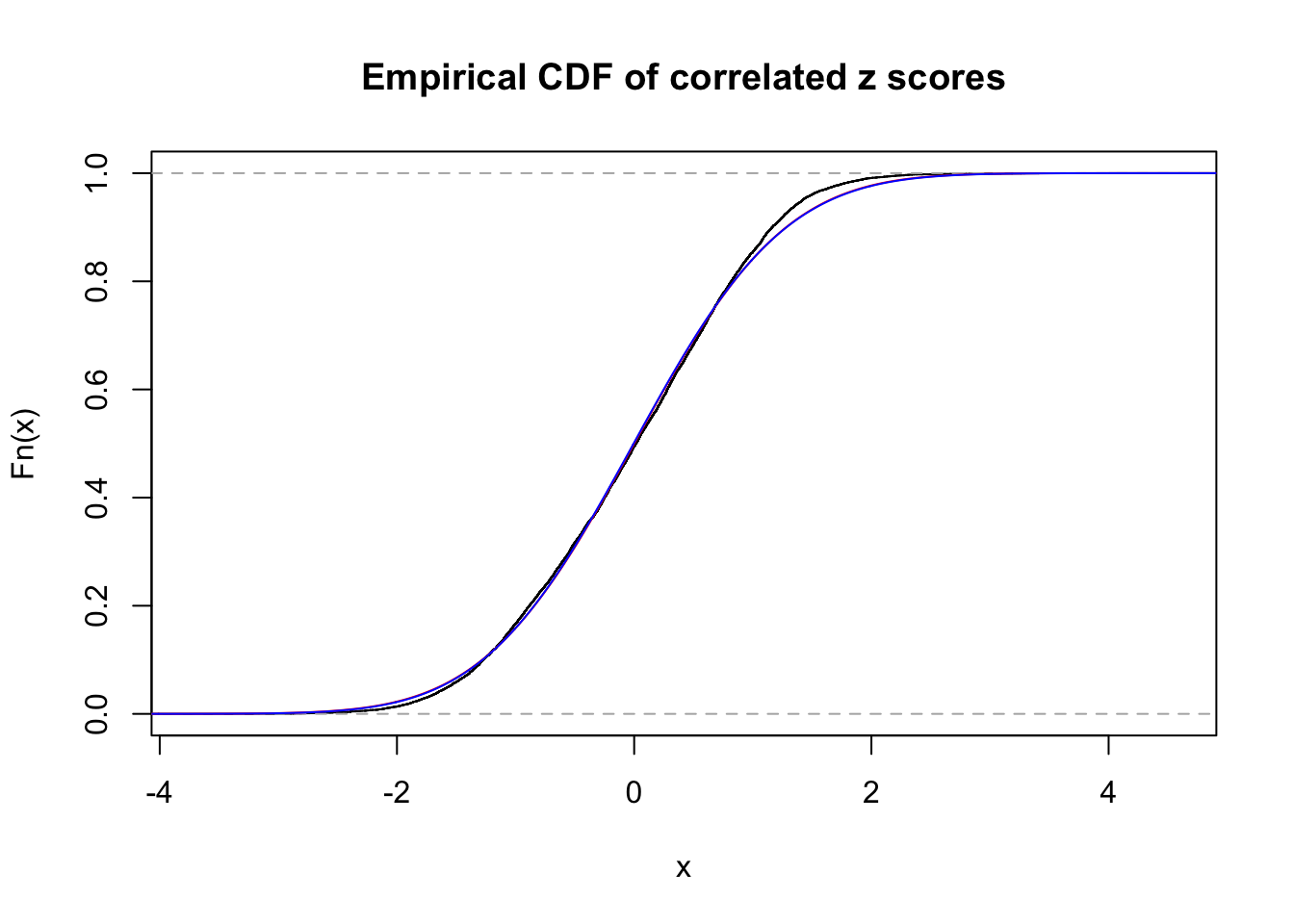
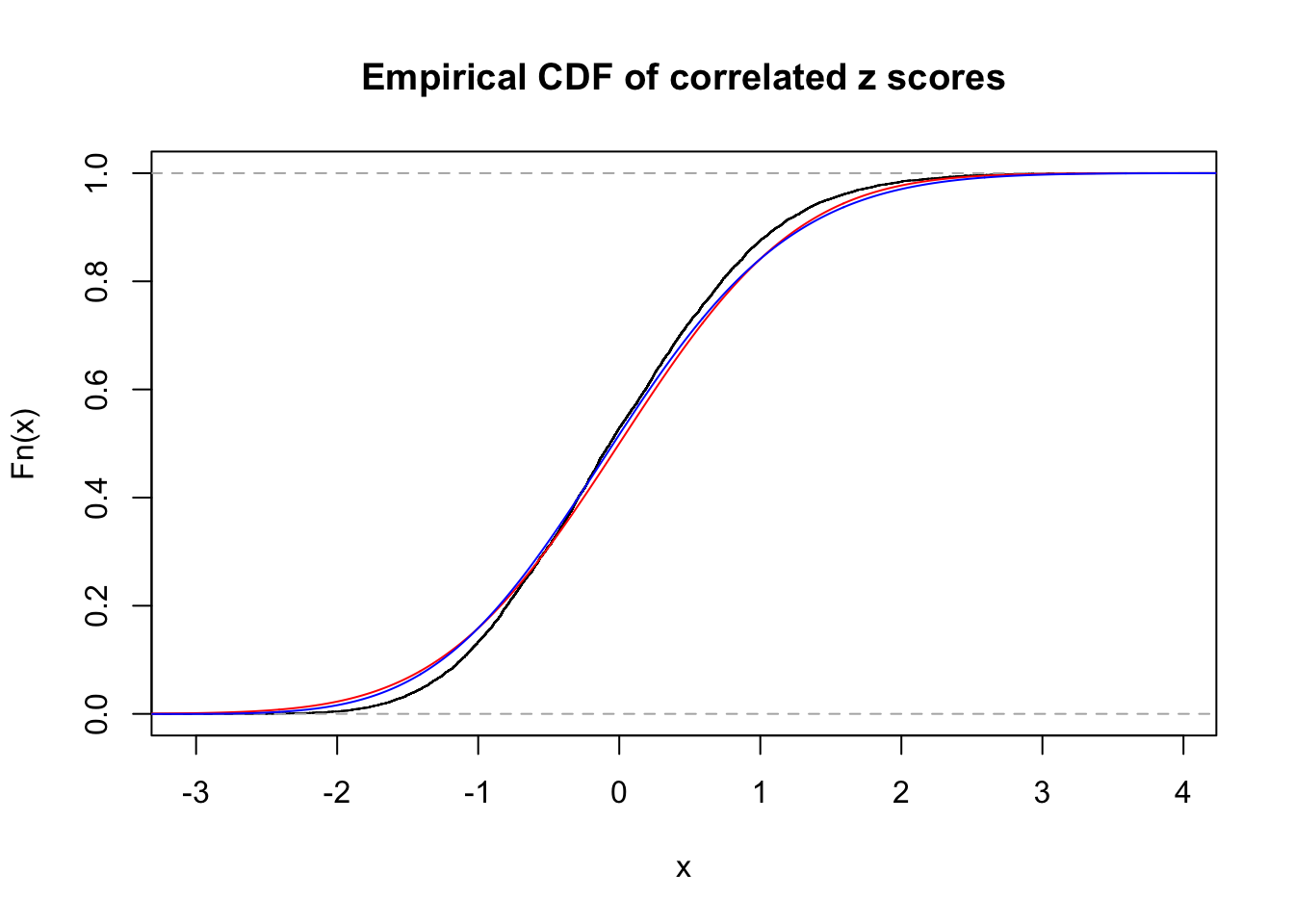
Data Set 34 :
Number of BH false discoveries: 0 ;
ASH's pihat0 = 1 ;
Normalized coefficients of Gaussian derivatives:
1 : 0 ; 2 : 0 ; 3 : 0.101371139384381 ; 4 : 0 ;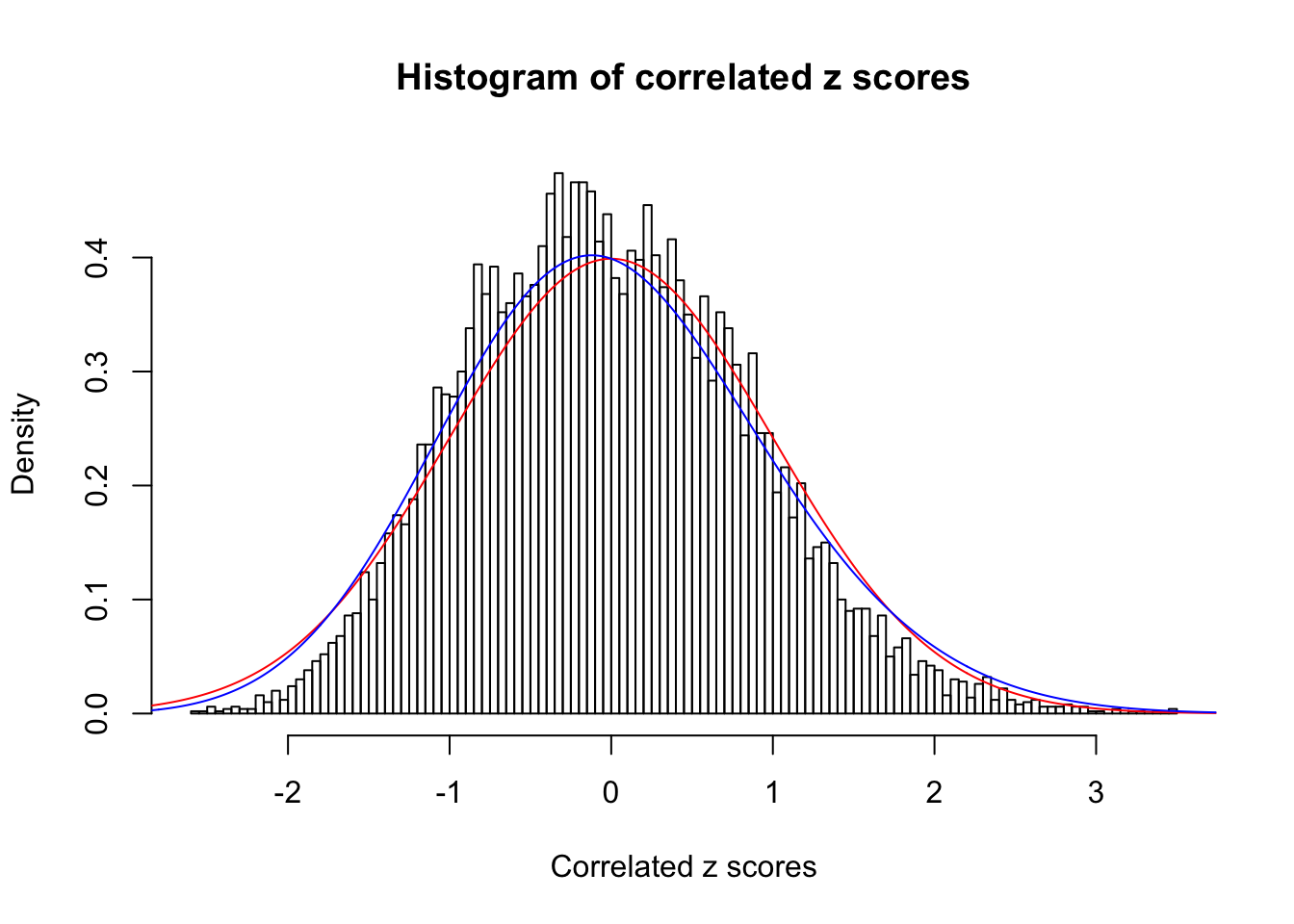
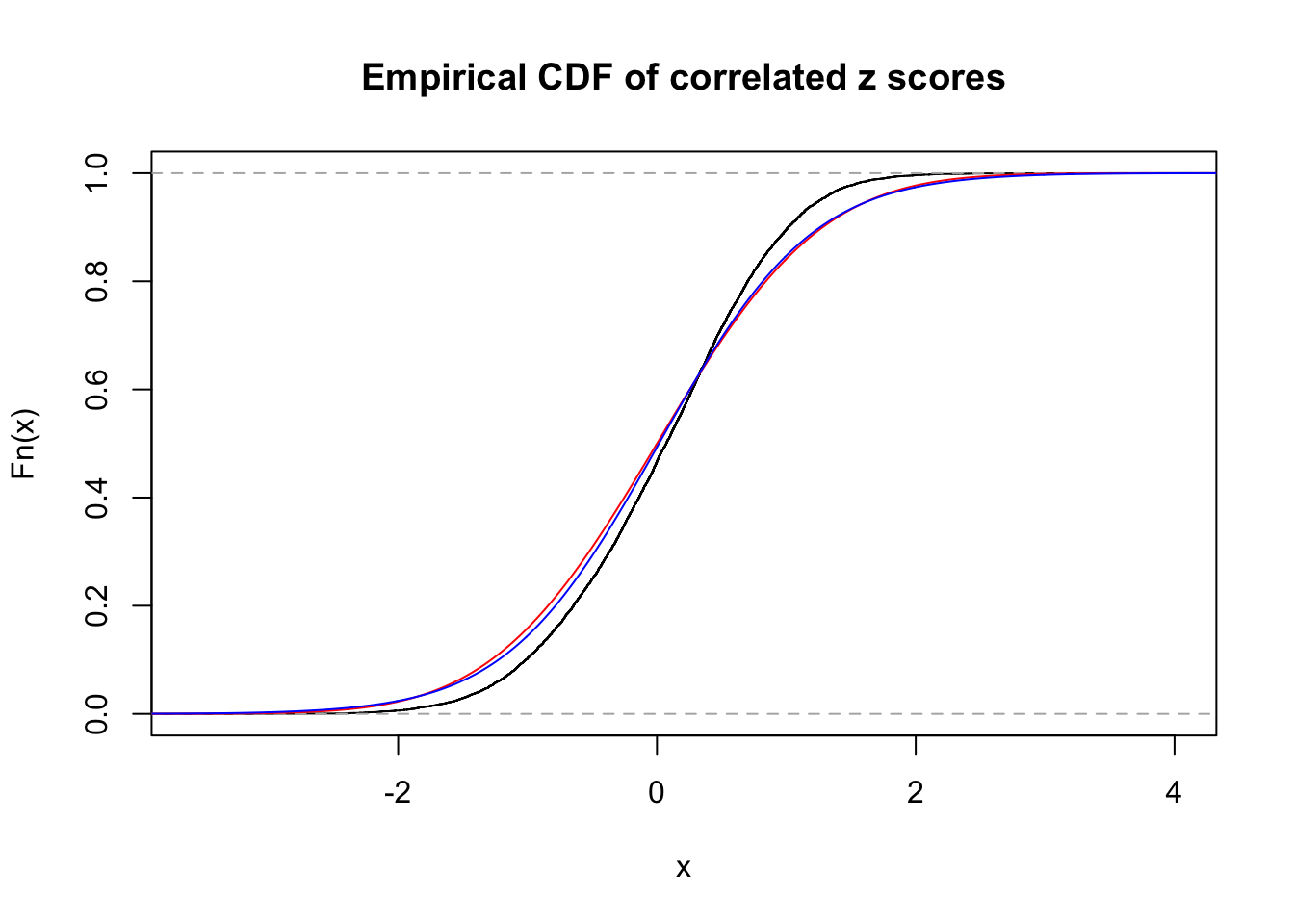
Data Set 35 :
Number of BH false discoveries: 0 ;
ASH's pihat0 = 1 ;
Normalized coefficients of Gaussian derivatives:
1 : 0.0165529780499778 ; 2 : 0 ; 3 : 0 ; 4 : 0.102360725261069 ;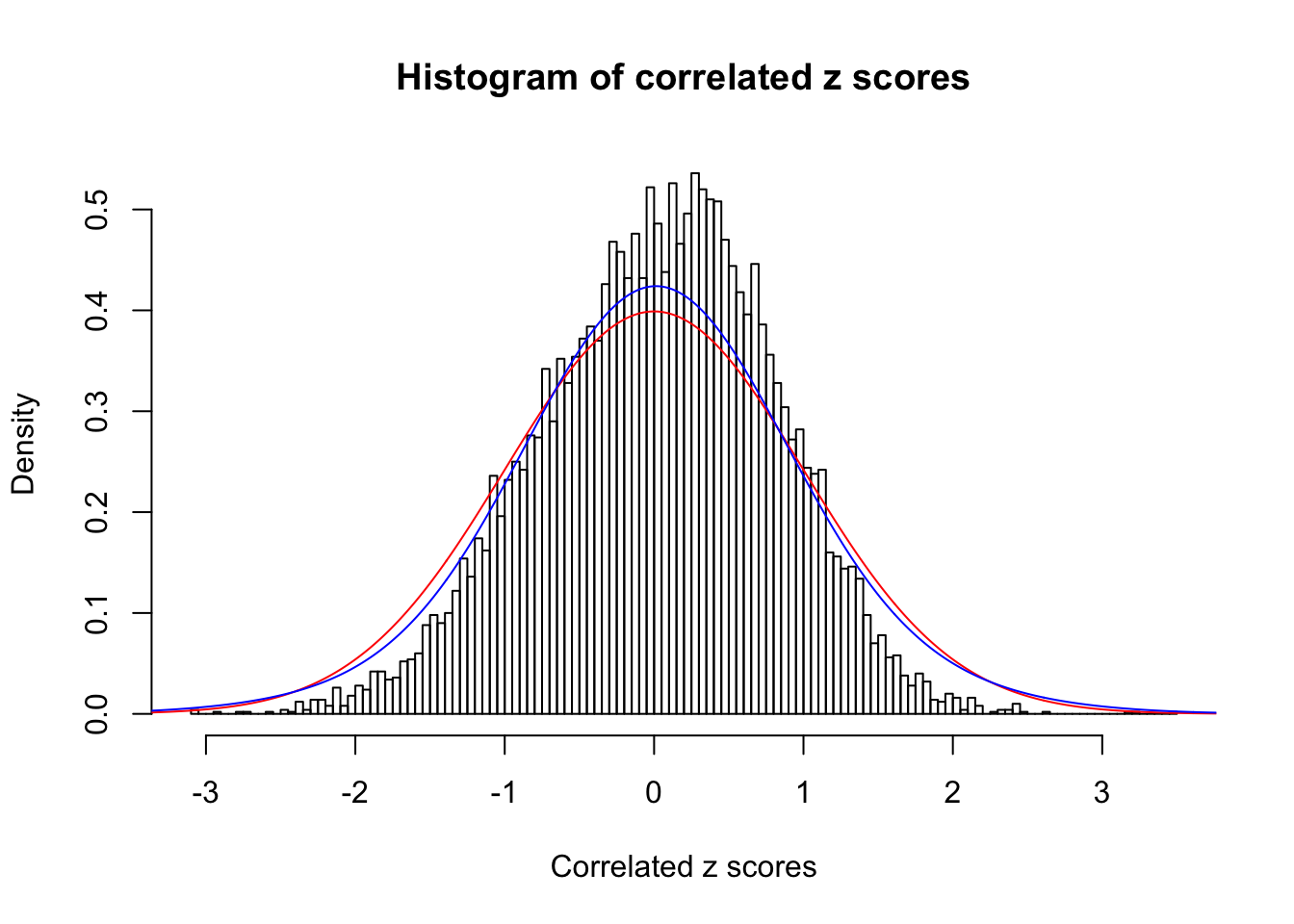
Data Set 36 :
Number of BH false discoveries: 0 ;
ASH's pihat0 = 1 ;
Normalized coefficients of Gaussian derivatives:
1 : 0.00780175544762885 ; 2 : 0 ; 3 : 0 ; 4 : 0.0653975820808355 ;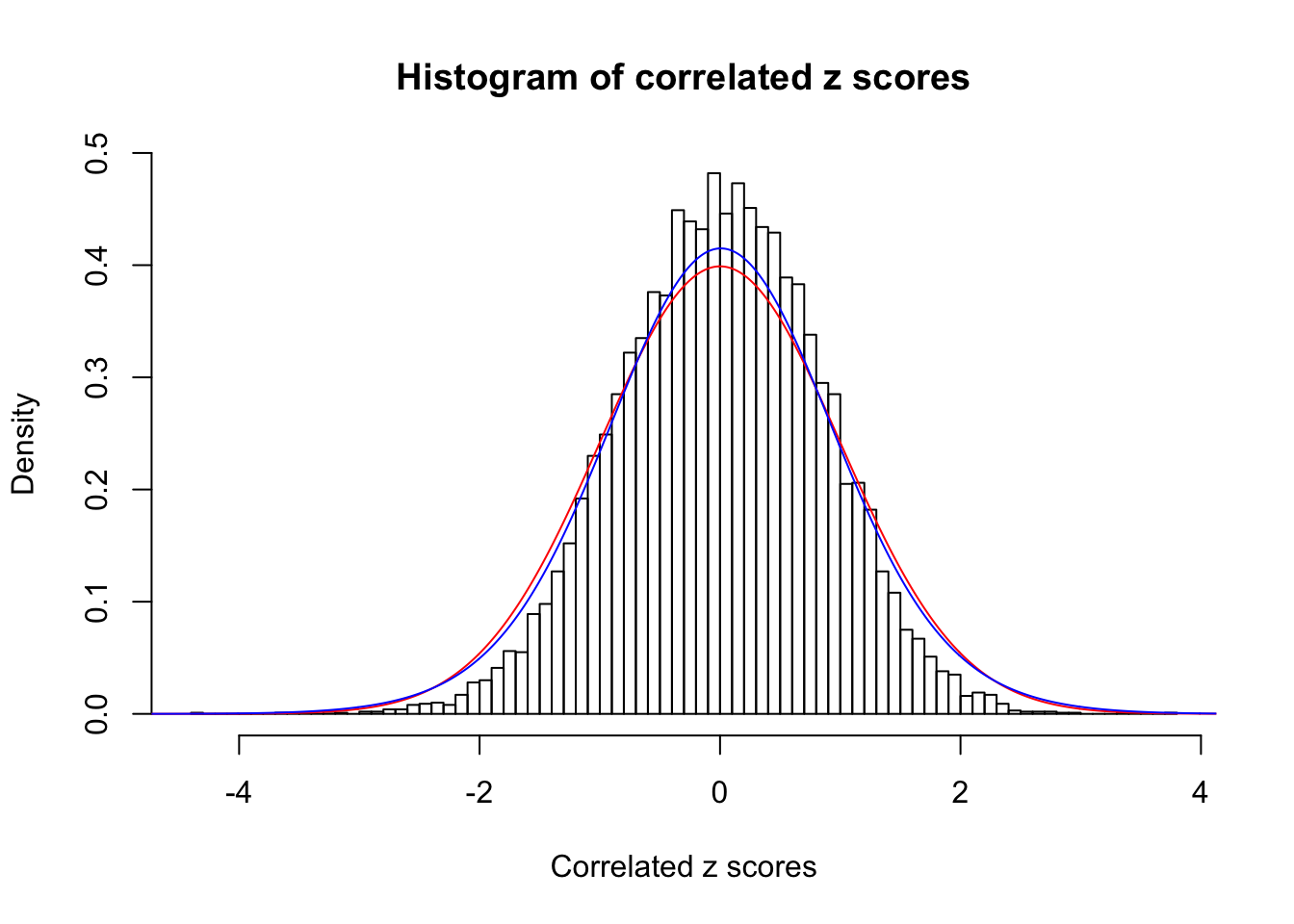
Data Set 38 :
Number of BH false discoveries: 0 ;
ASH's pihat0 = 1 ;
Normalized coefficients of Gaussian derivatives:
1 : 0 ; 2 : 0 ; 3 : 0 ; 4 : 0.0643337534695275 ;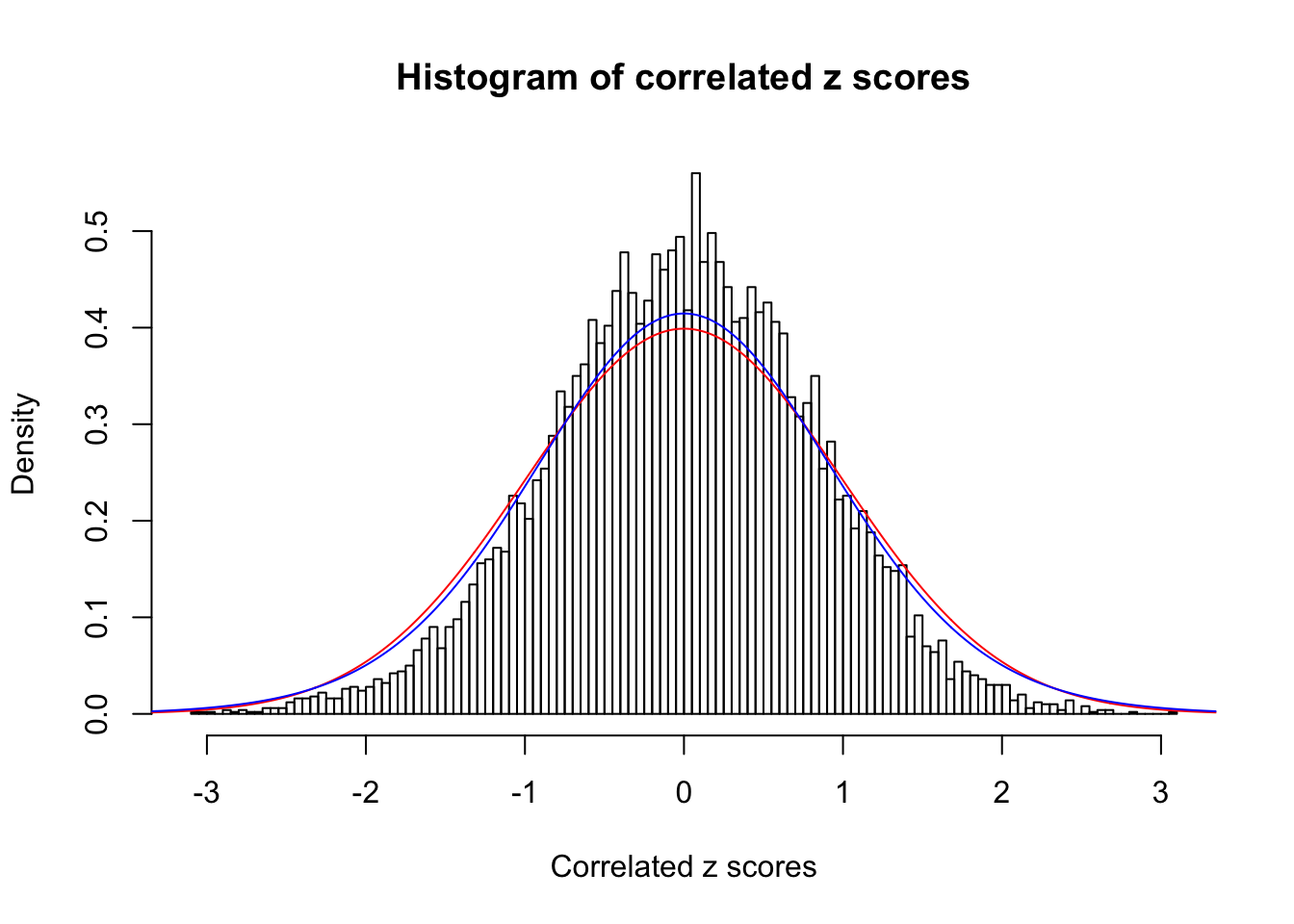
Data Set 42 :
Number of BH false discoveries: 0 ;
ASH's pihat0 = 1 ;
Normalized coefficients of Gaussian derivatives:
1 : 0 ; 2 : 0 ; 3 : 0.0117929353952078 ; 4 : 0.14259210767113 ;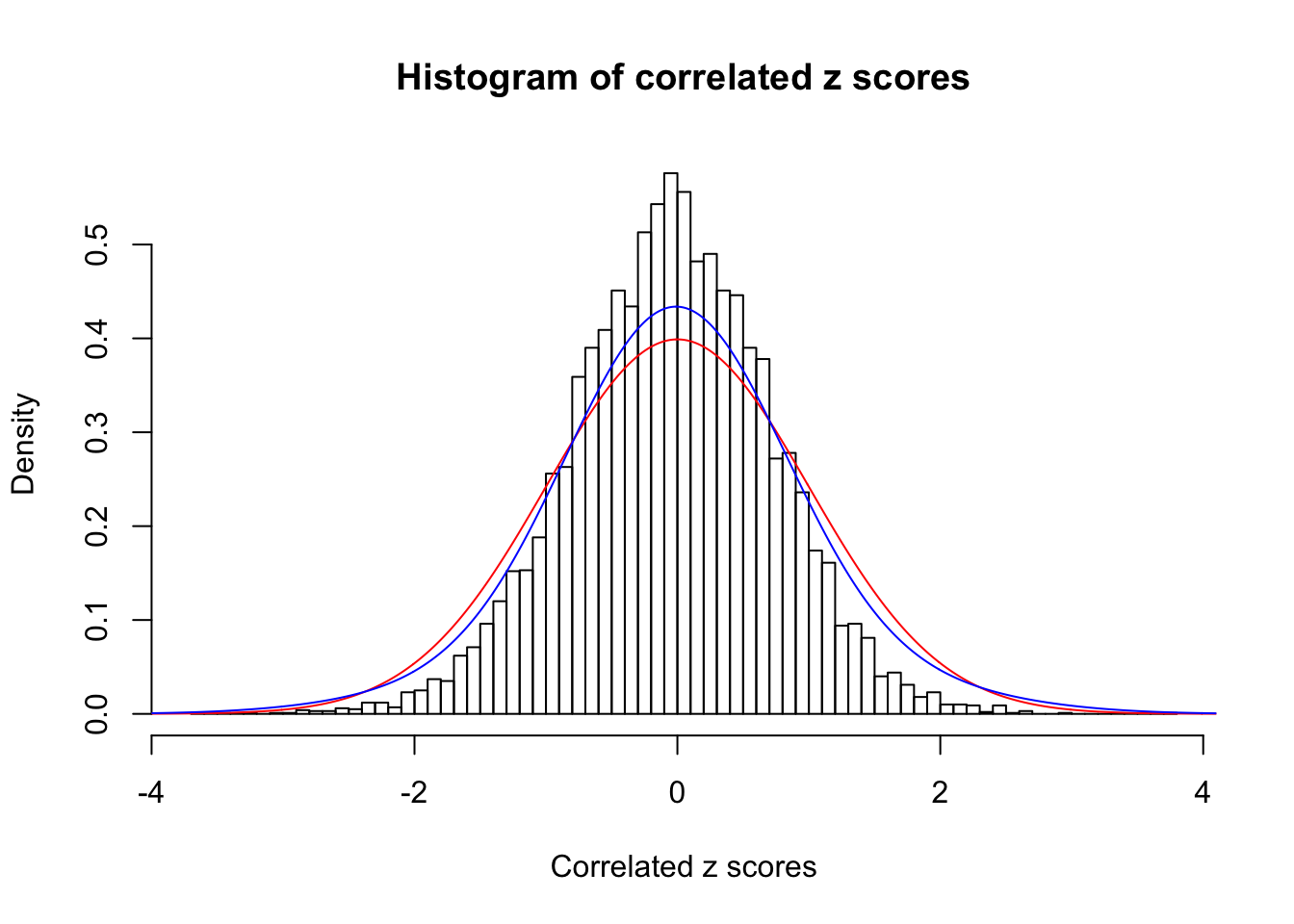
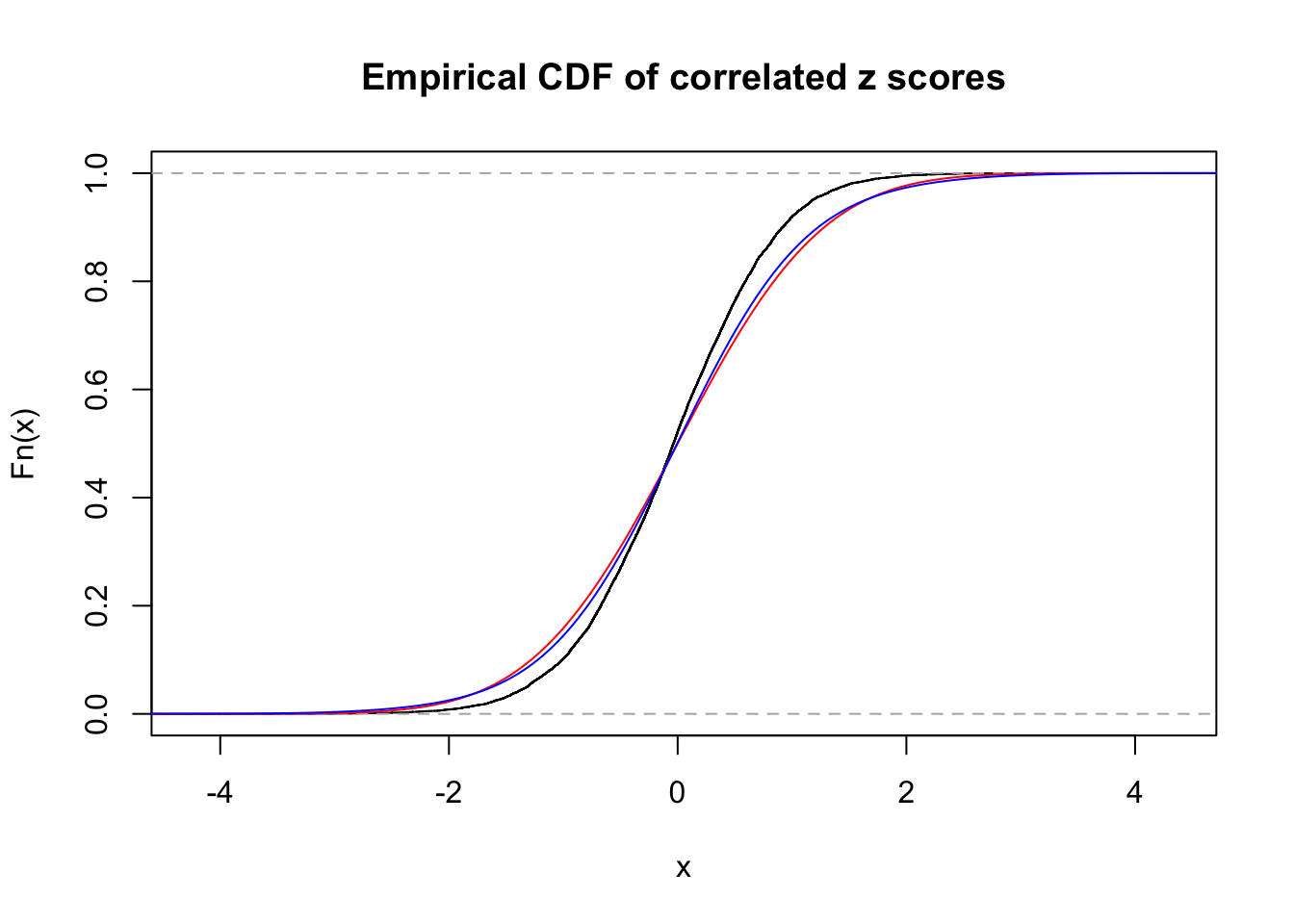
Data Set 43 :
Number of BH false discoveries: 0 ;
ASH's pihat0 = 1 ;
Normalized coefficients of Gaussian derivatives:
1 : 0 ; 2 : 0 ; 3 : 0.0266591670097924 ; 4 : 0.119934975949713 ;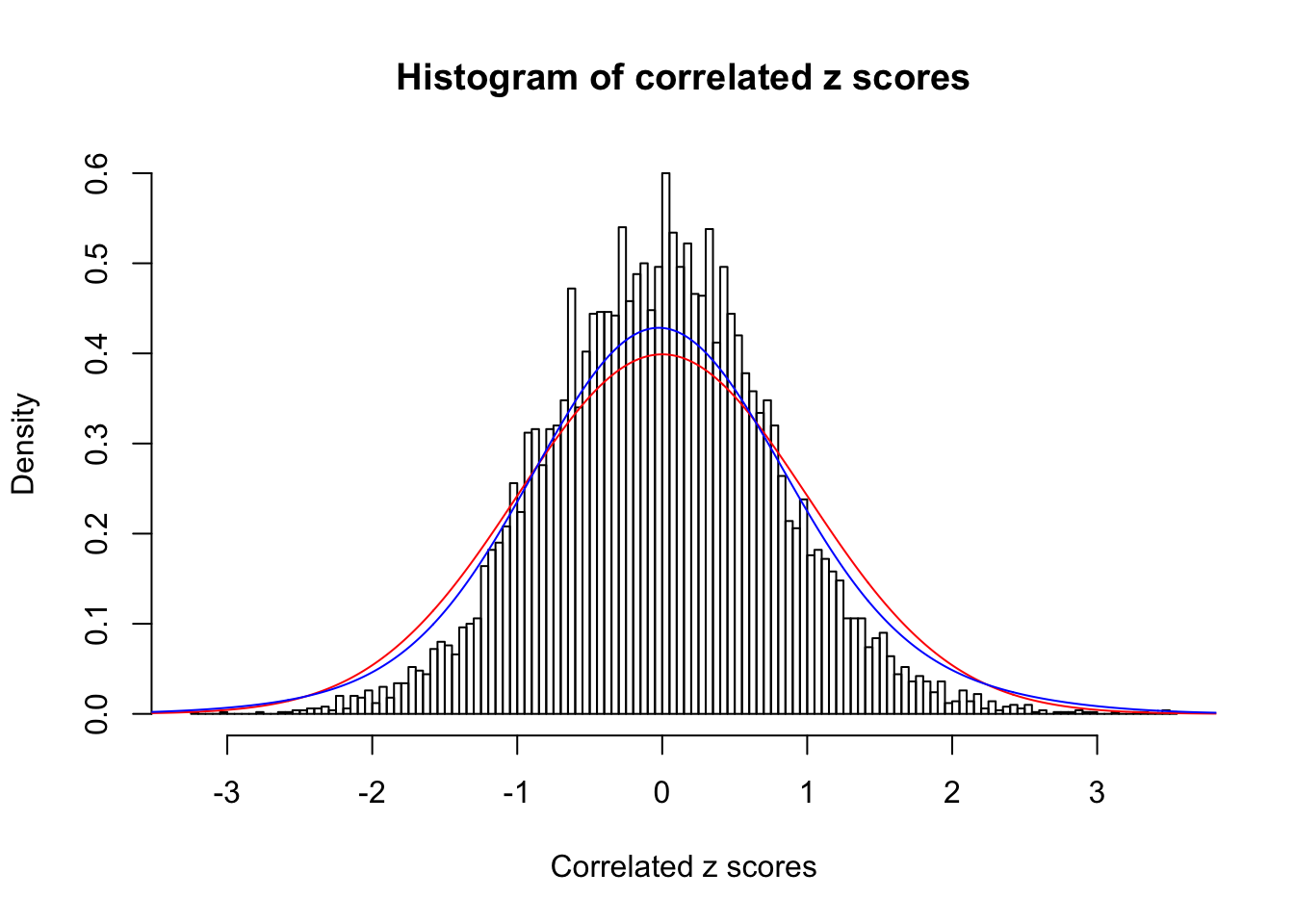
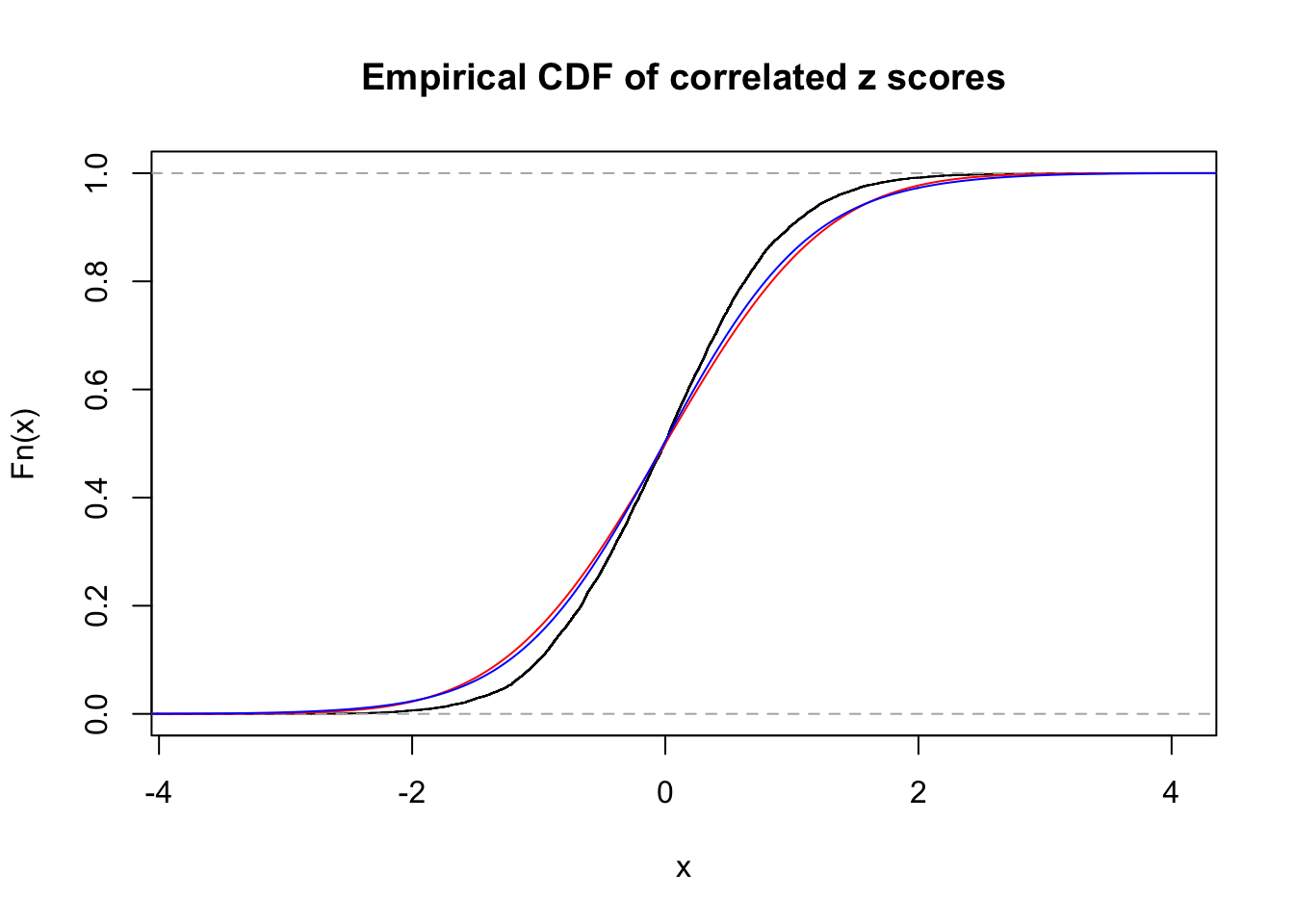
Data Set 45 :
Number of BH false discoveries: 0 ;
ASH's pihat0 = 1 ;
Normalized coefficients of Gaussian derivatives:
1 : 0.0197168202529947 ; 2 : 0 ; 3 : 0.00535774384797348 ; 4 : 0.0981326520063197 ;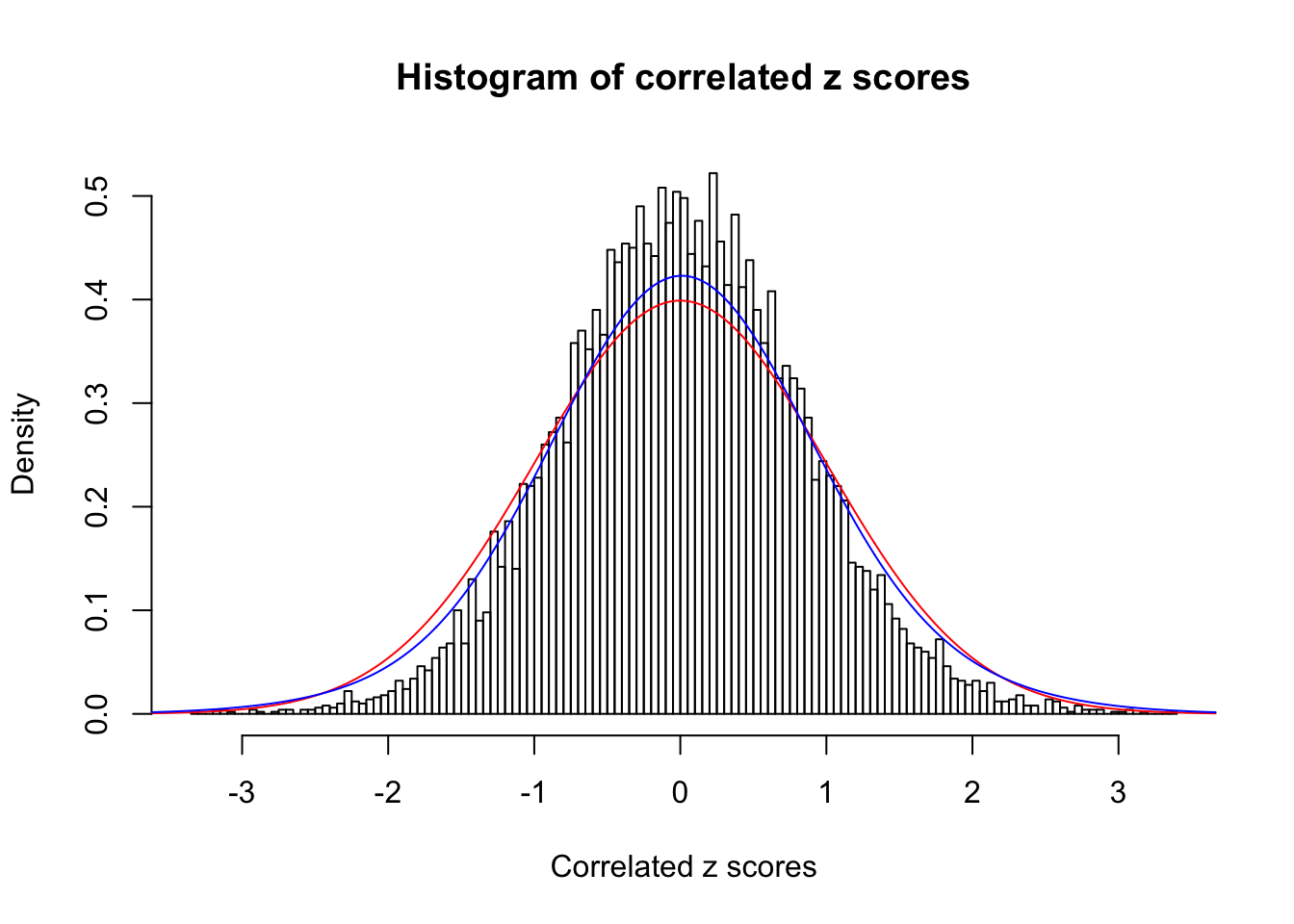
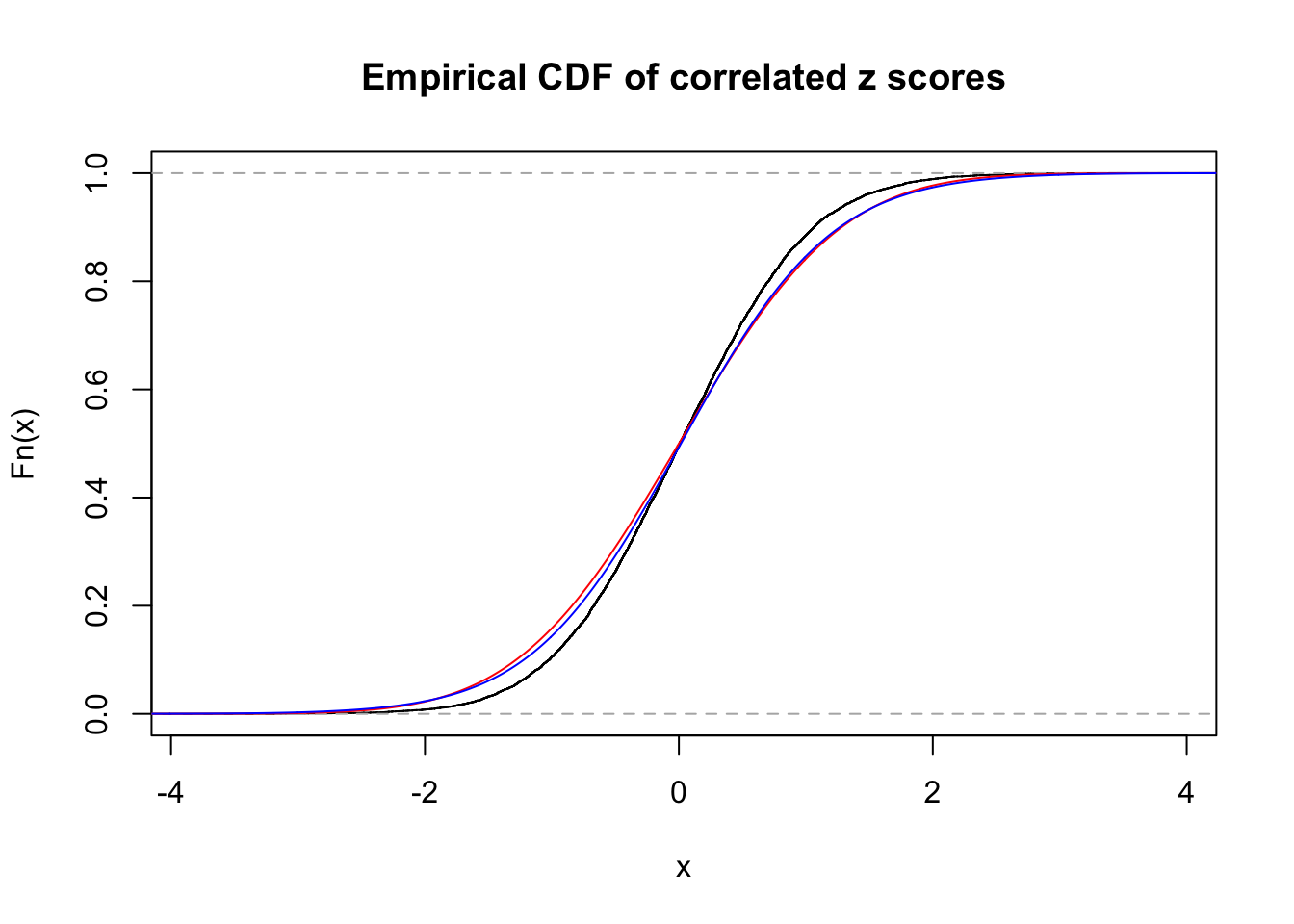
Data Set 46 :
Number of BH false discoveries: 0 ;
ASH's pihat0 = 1 ;
Normalized coefficients of Gaussian derivatives:
1 : 0 ; 2 : 0 ; 3 : 0.0142593356797949 ; 4 : 0 ;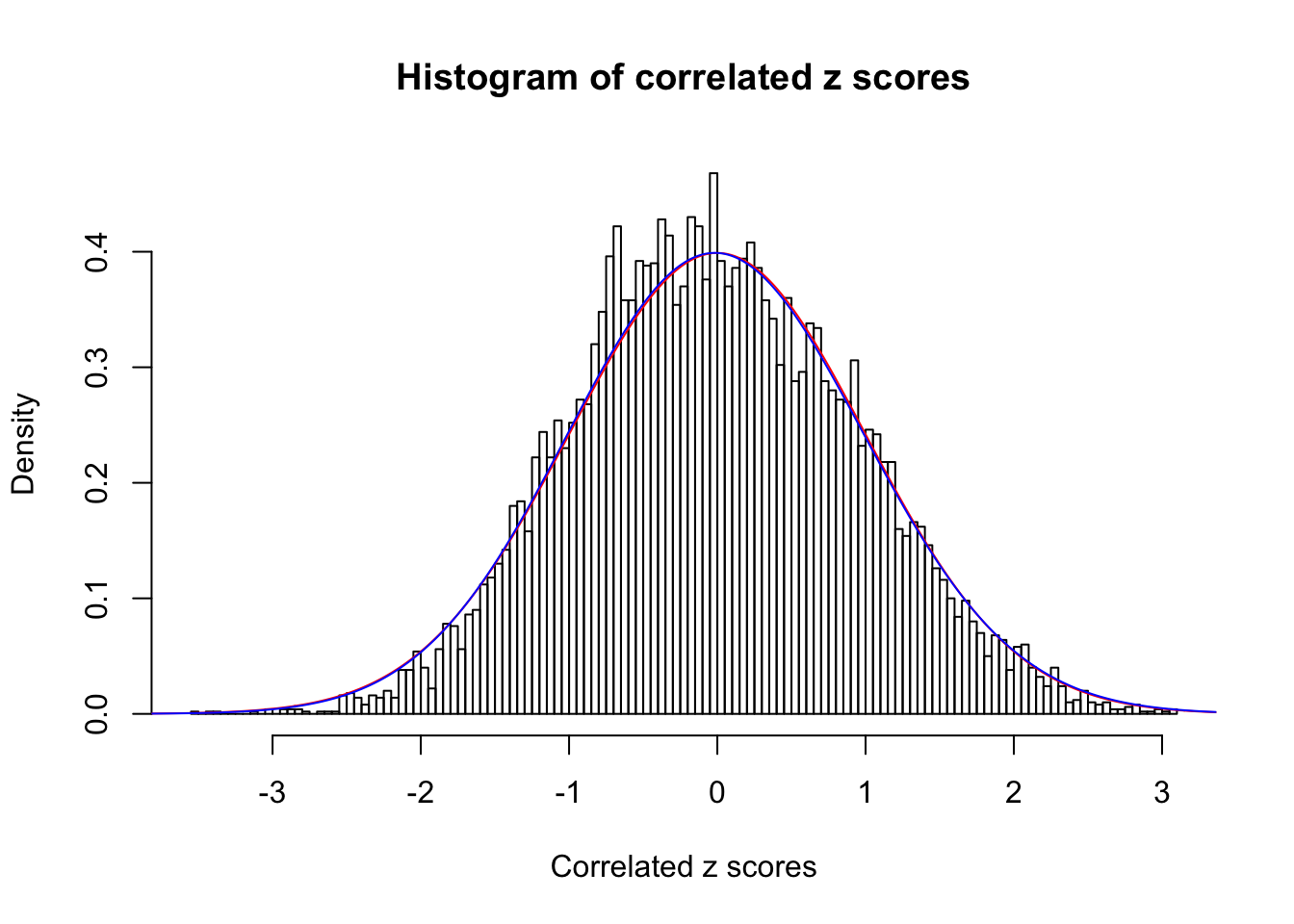
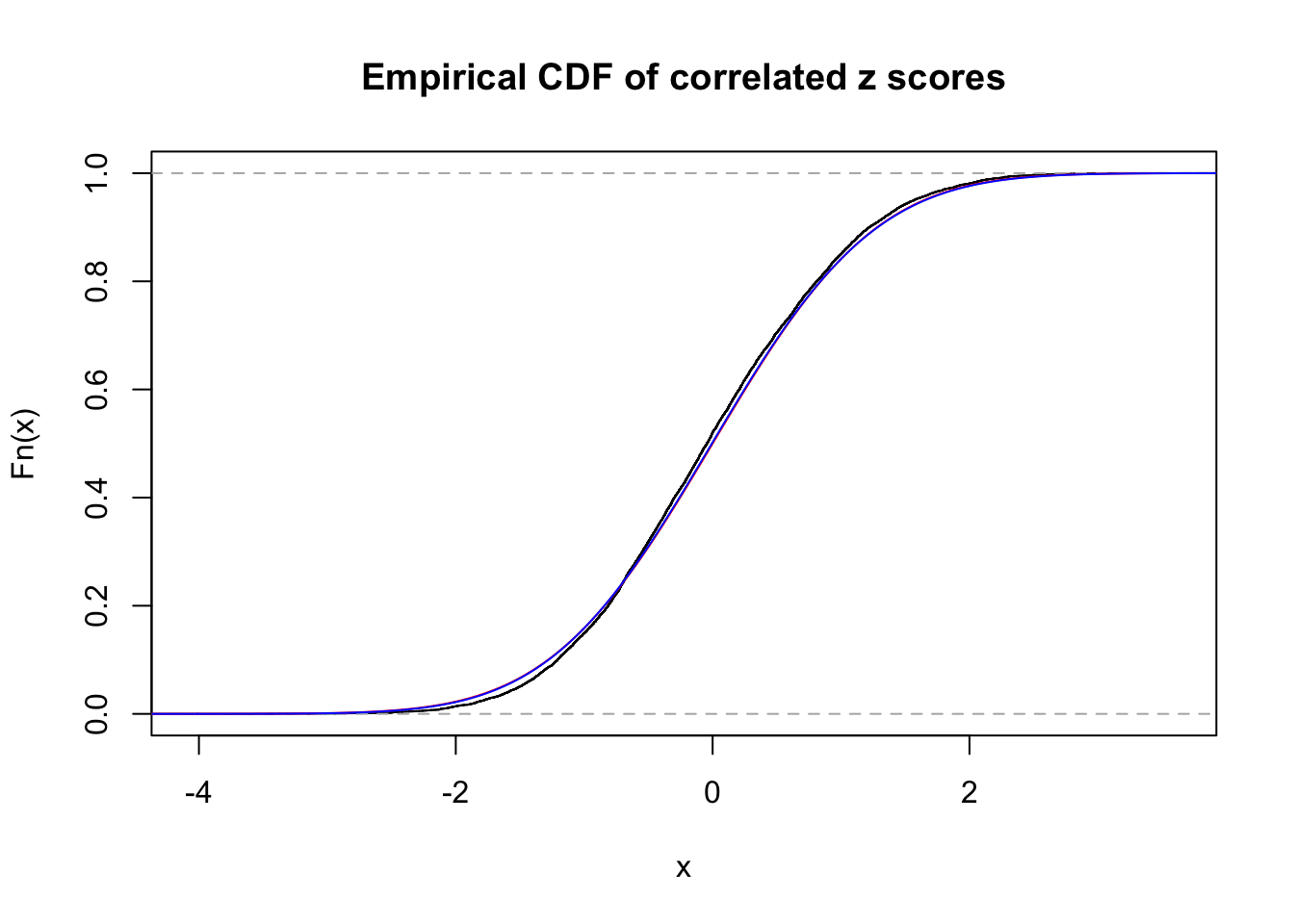
Data Set 47 :
Number of BH false discoveries: 0 ;
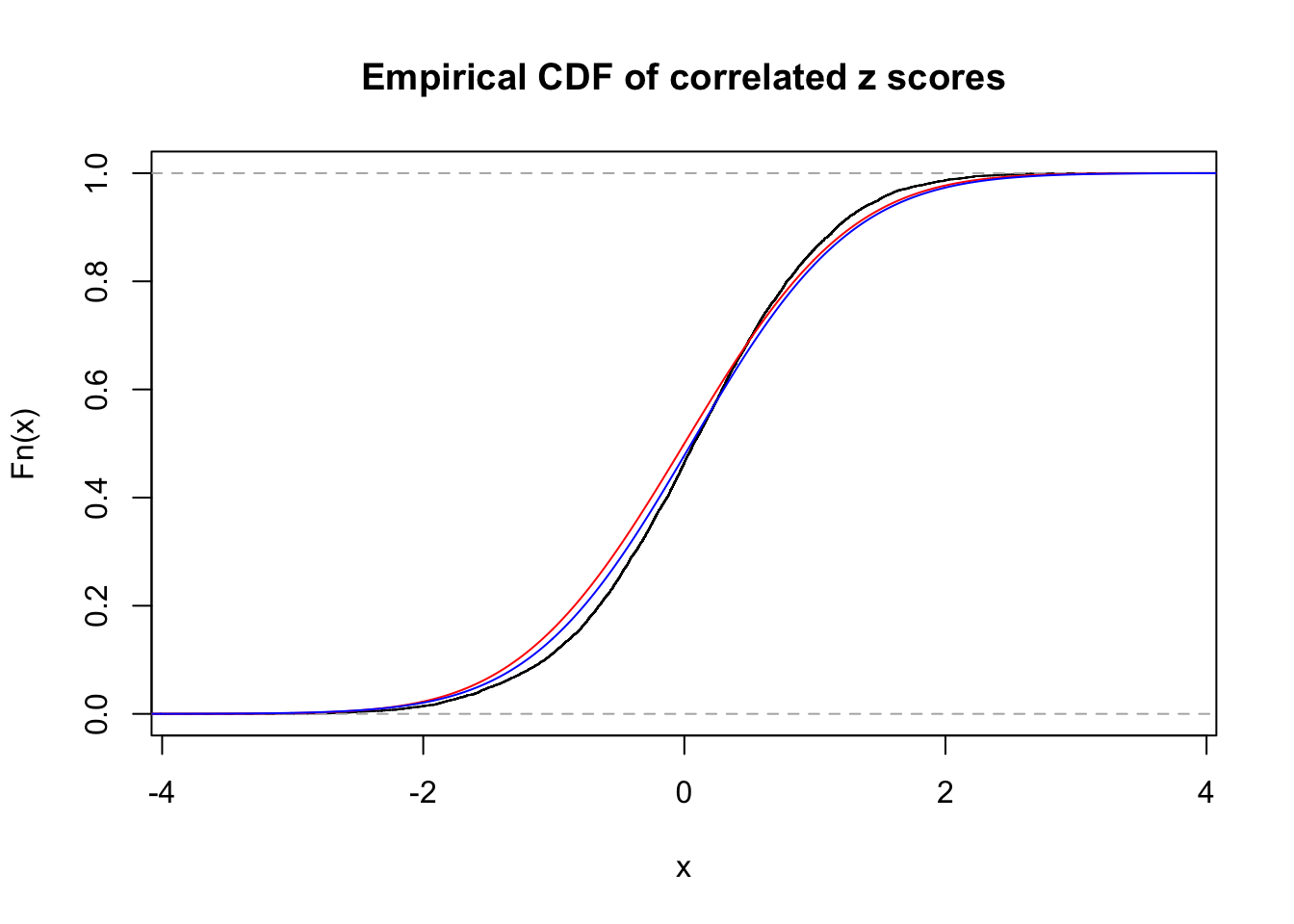
ASH's pihat0 = 1 ;
Normalized coefficients of Gaussian derivatives:
1 : 0.0553713103730739 ; 2 : 0 ; 3 : 0 ; 4 : 0.0424140129177028 ;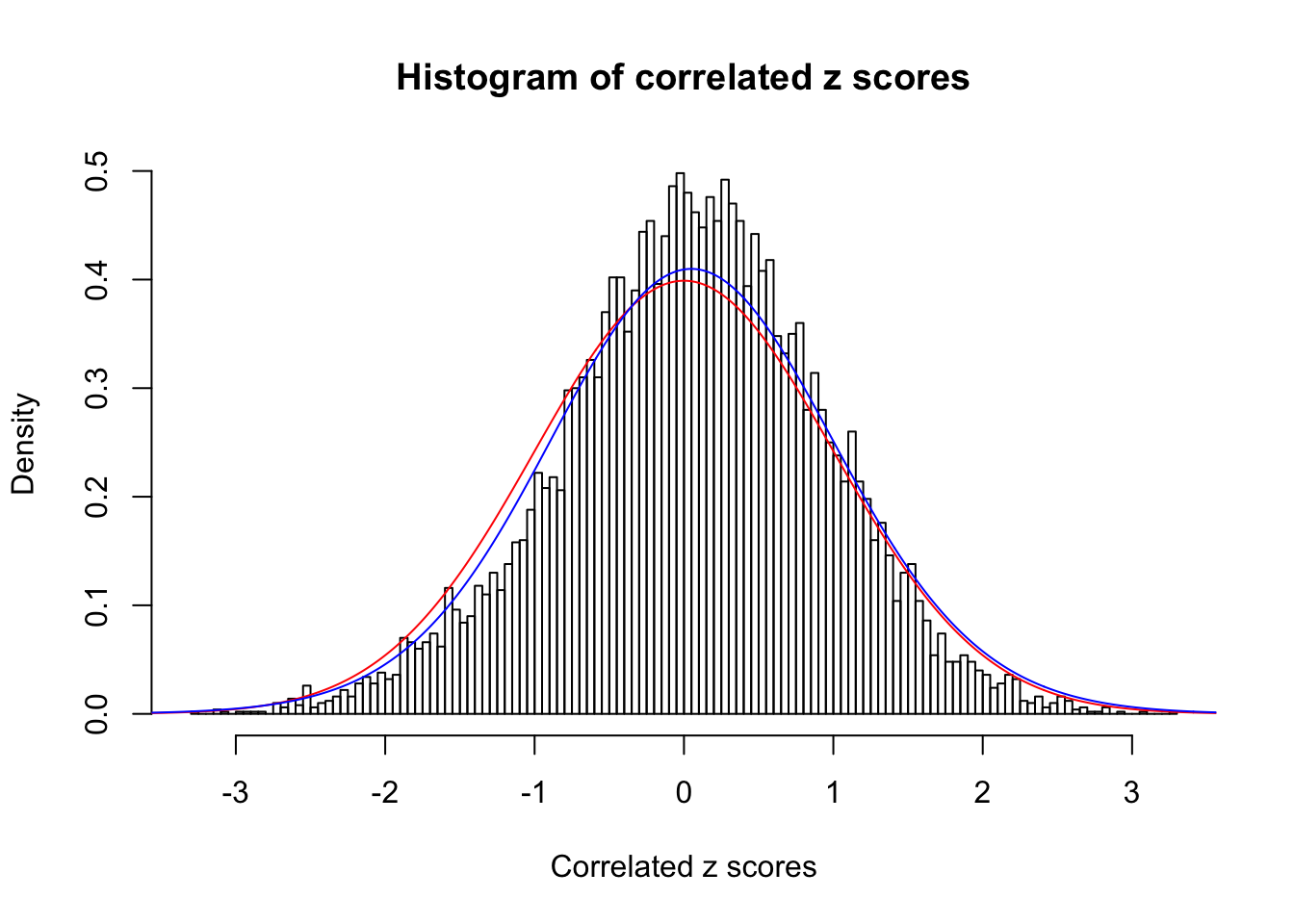
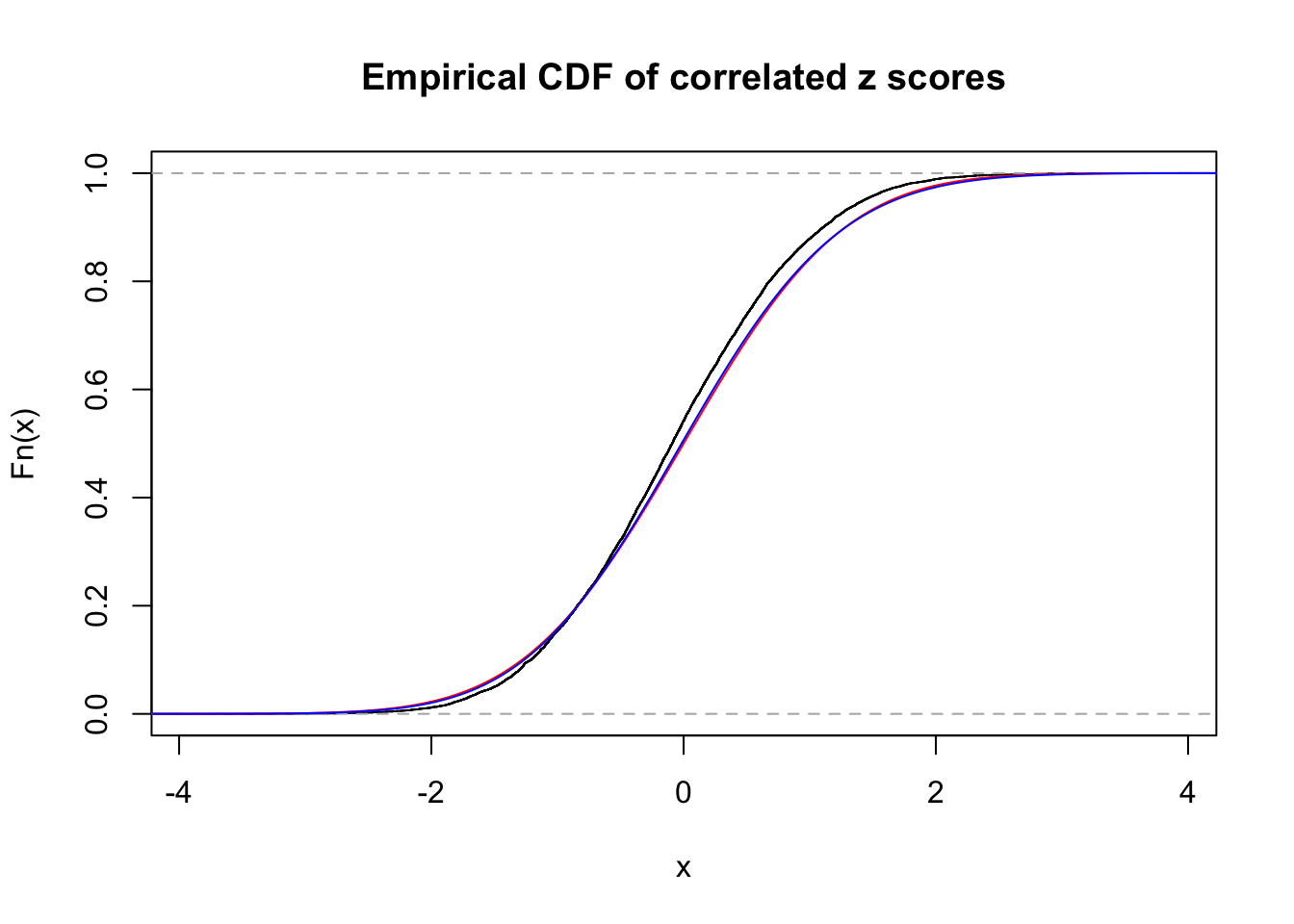
Data Set 49 :
Number of BH false discoveries: 0 ;
ASH's pihat0 = 1 ;
Normalized coefficients of Gaussian derivatives:
1 : 0 ; 2 : 0 ; 3 : 0.0399557075219927 ; 4 : 0.0203389185614439 ;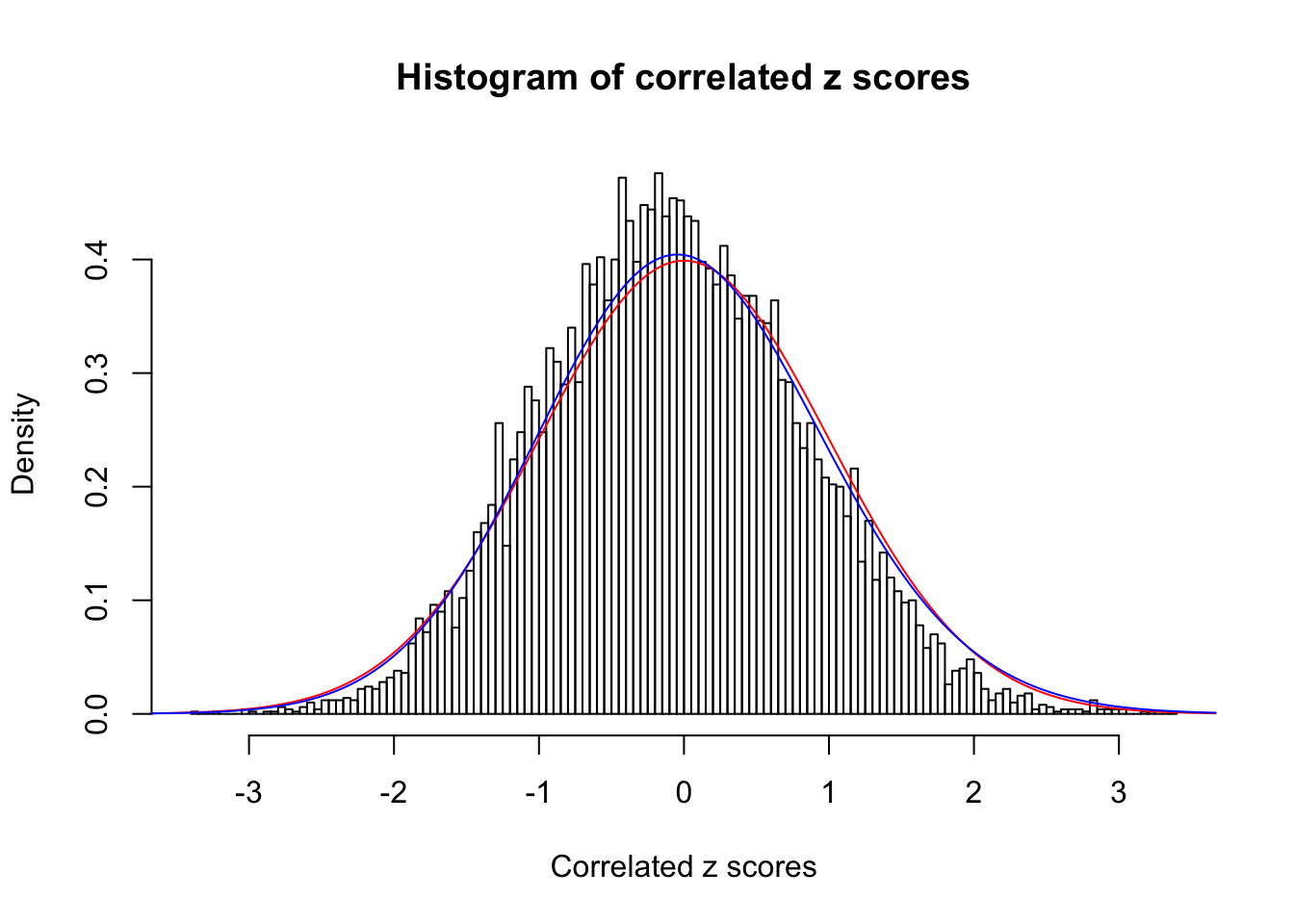
Data Set 52 :
Number of BH false discoveries: 0 ;
ASH's pihat0 = 1 ;
Normalized coefficients of Gaussian derivatives:
1 : 0.0717118497321021 ; 2 : 0 ; 3 : 0 ; 4 : 0 ;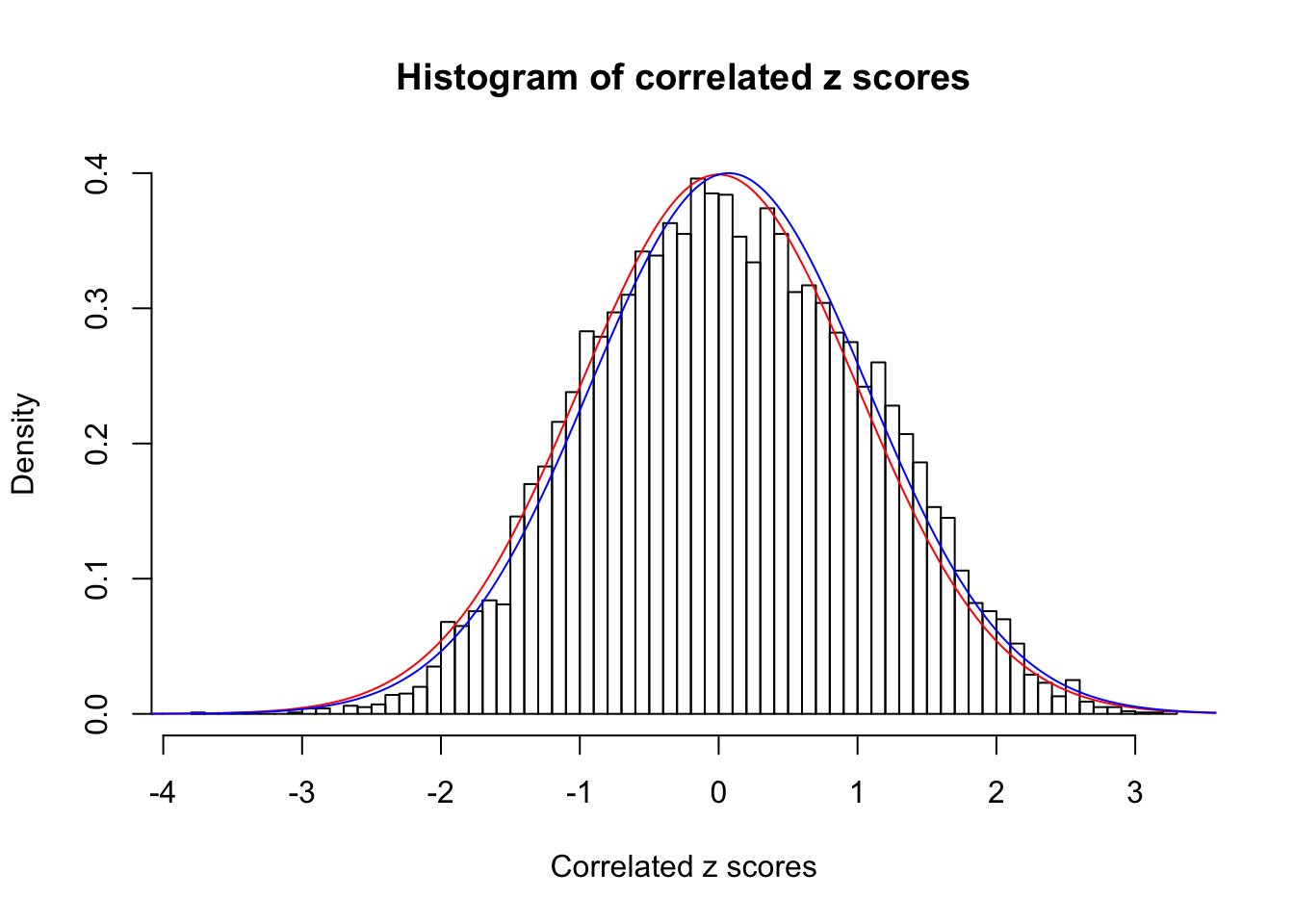
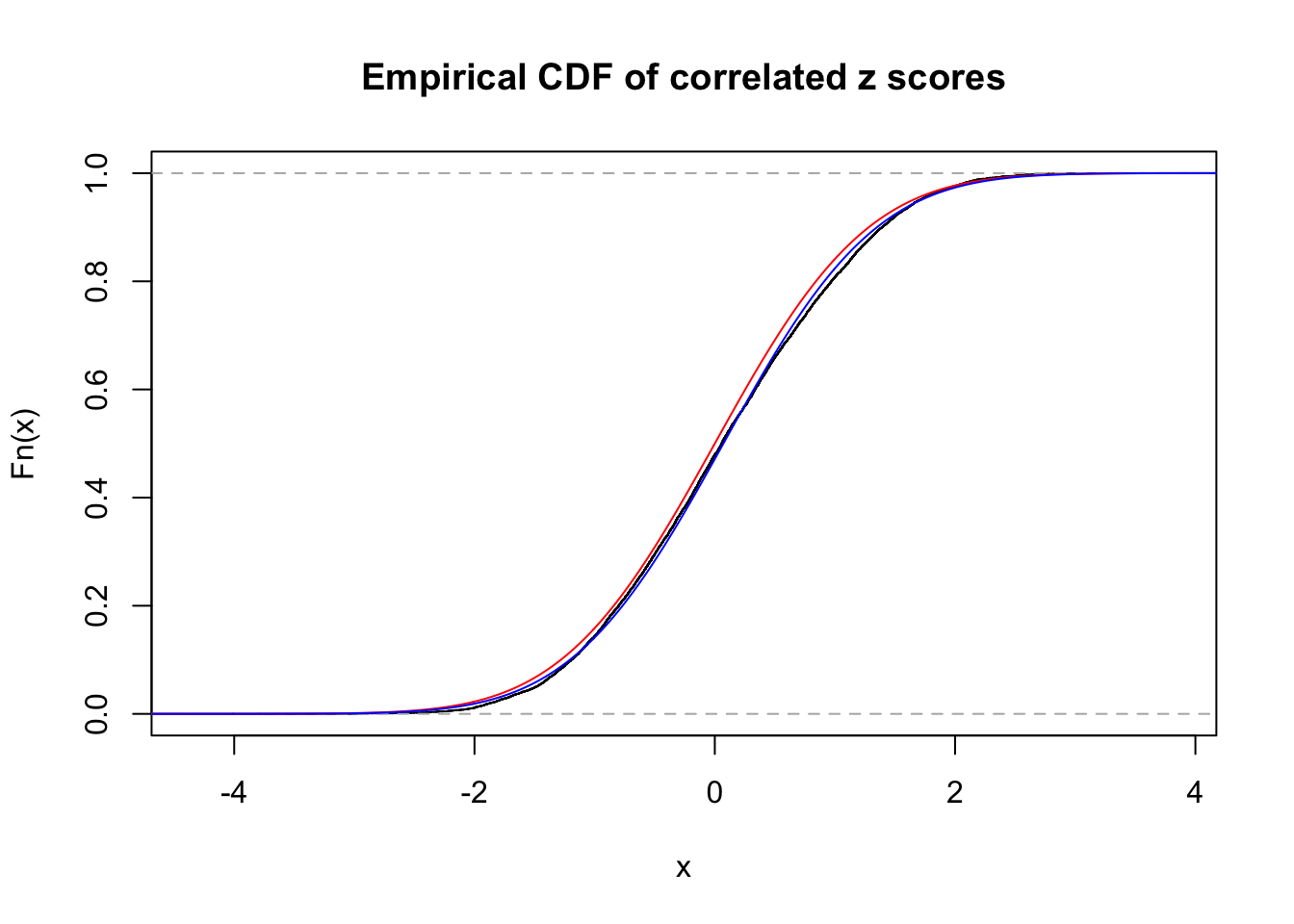
Data Set 53 :
Number of BH false discoveries: 0 ;
ASH's pihat0 = 1 ;
Normalized coefficients of Gaussian derivatives:
1 : 0 ; 2 : 0 ; 3 : 0.0203994359033729 ; 4 : 0 ;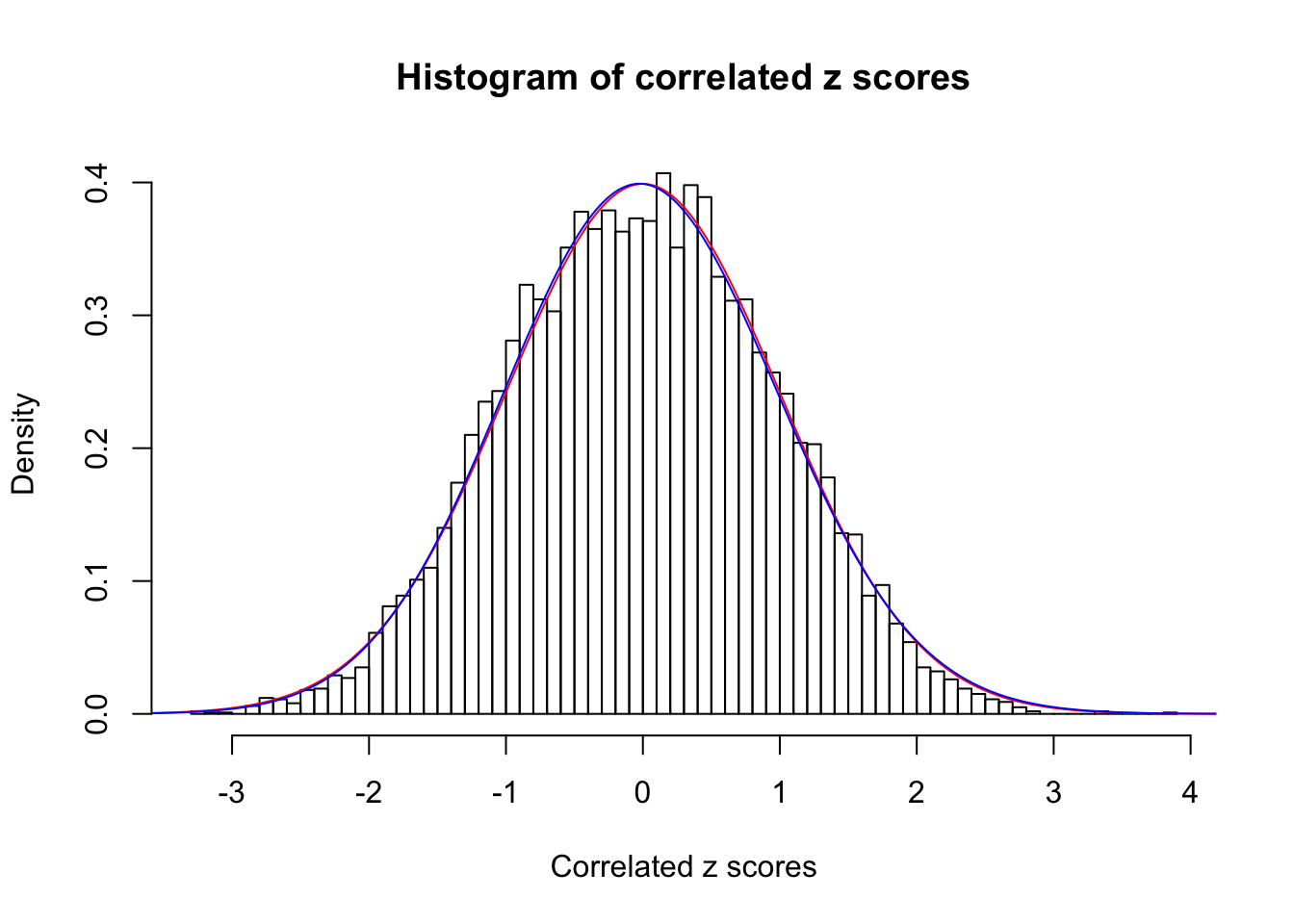
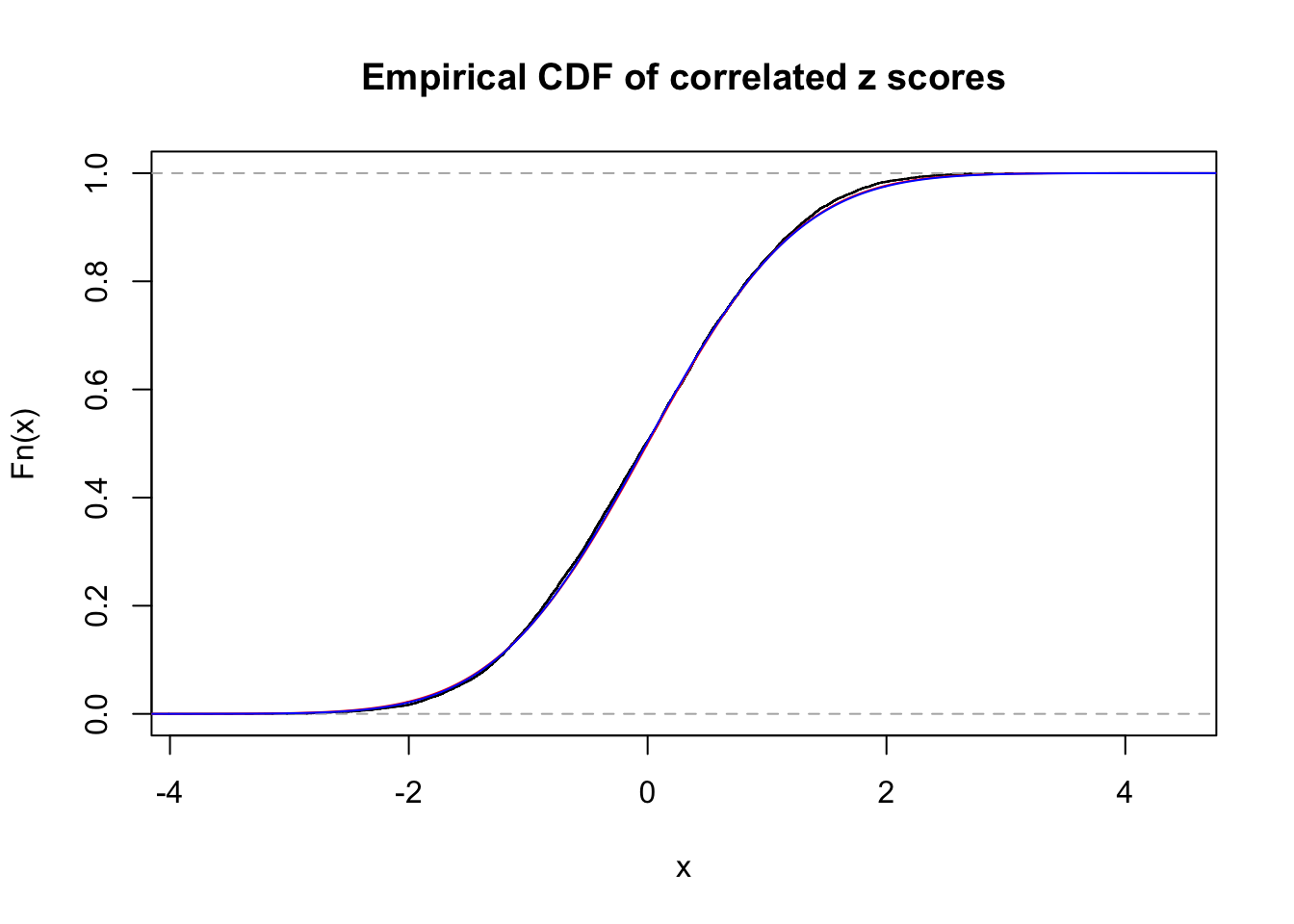
Data Set 54 :
Number of BH false discoveries: 0 ;
ASH's pihat0 = 1 ;
Normalized coefficients of Gaussian derivatives:
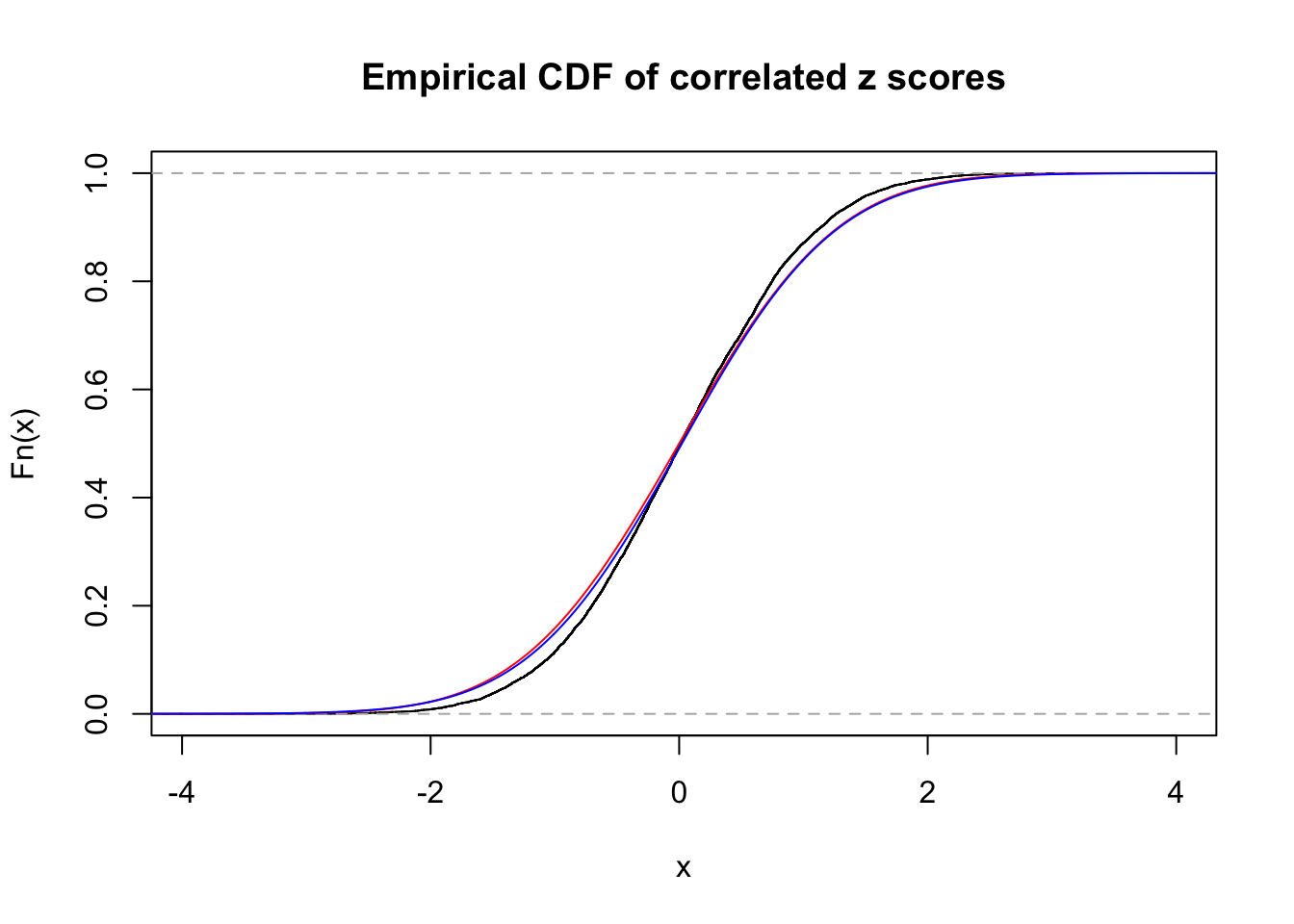
1 : 0.0226491329325567 ; 2 : 0 ; 3 : 0 ; 4 : 0.0369913458520336 ;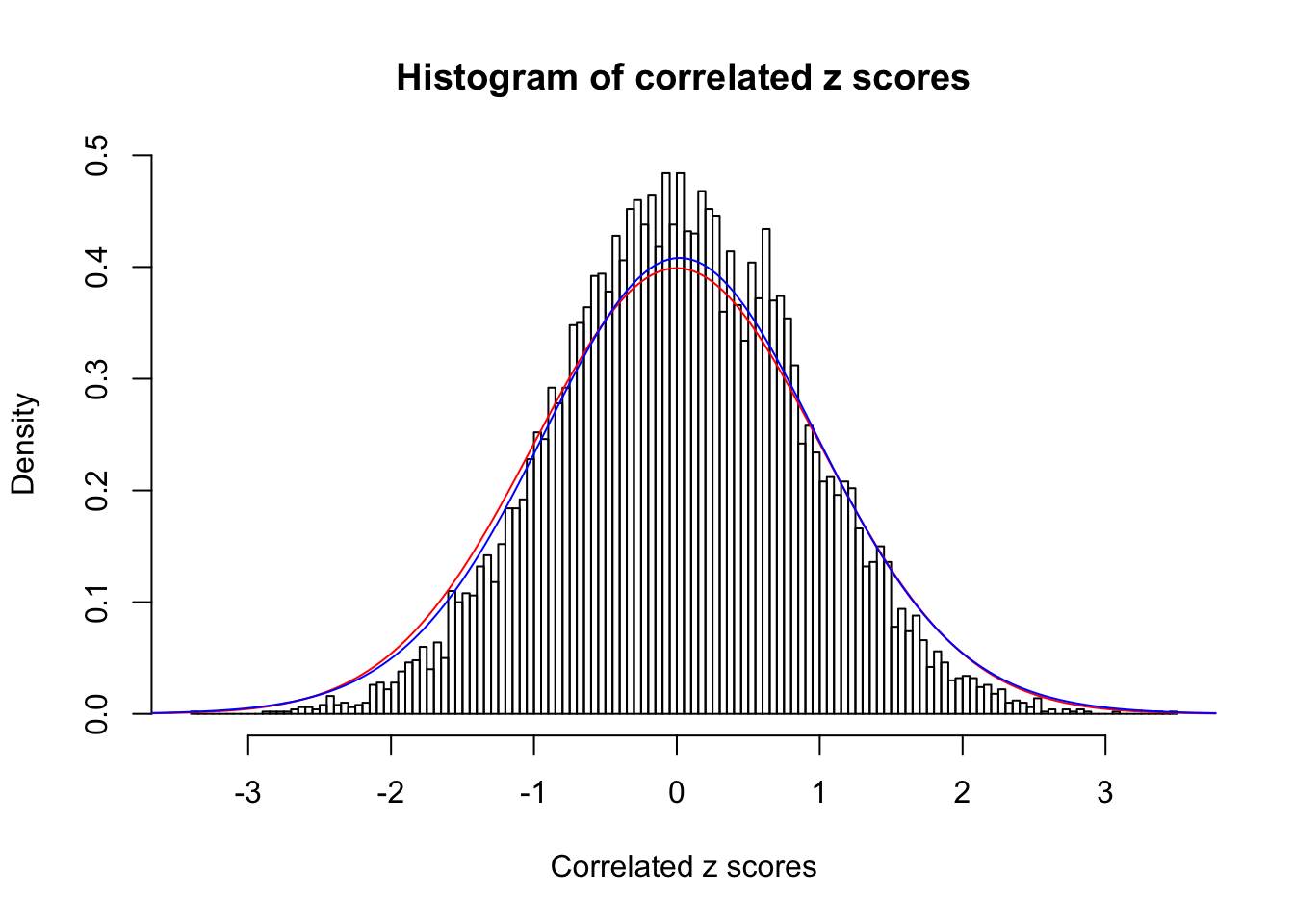
Remarks
The implementation by Rmosek is more convenient, cleaner, yet more complicated. Need to understand different issues such as initialization, optimal orders, controlling parameters, dual construction, etc.
Session information
sessionInfo()R version 3.4.2 (2017-09-28)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS Sierra 10.12.6
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] Rmosek_7.1.3 PolynomF_0.94 cvxr_0.0.0.9400 REBayes_0.85
[5] Matrix_1.2-11 SQUAREM_2017.10-1 EQL_1.0-0 ttutils_1.0-1
loaded via a namespace (and not attached):
[1] Rcpp_0.12.13 lattice_0.20-35 gmp_0.5-13.1 digest_0.6.12
[5] rprojroot_1.2 MASS_7.3-47 grid_3.4.2 backports_1.1.1
[9] git2r_0.19.0 magrittr_1.5 evaluate_0.10.1 stringi_1.1.5
[13] rmarkdown_1.6 tools_3.4.2 stringr_1.2.0 yaml_2.1.14
[17] compiler_3.4.2 htmltools_0.3.6 knitr_1.17 This R Markdown site was created with workflowr