Variability of Knockoff’s FDR Control
Lei Sun
2018-04-03
Last updated: 2018-05-15
workflowr checks: (Click a bullet for more information)-
✔ R Markdown file: up-to-date
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
-
✔ Environment: empty
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
-
✔ Seed:
set.seed(12345)The command
set.seed(12345)was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible. -
✔ Session information: recorded
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
-
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.✔ Repository version: 388e65e
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can usewflow_publishorwflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.Ignored files: Ignored: .DS_Store Ignored: .Rhistory Ignored: .Rproj.user/ Ignored: analysis/.DS_Store Ignored: analysis/BH_robustness_cache/ Ignored: analysis/FDR_Null_cache/ Ignored: analysis/FDR_null_betahat_cache/ Ignored: analysis/Rmosek_cache/ Ignored: analysis/StepDown_cache/ Ignored: analysis/alternative2_cache/ Ignored: analysis/alternative_cache/ Ignored: analysis/ash_gd_cache/ Ignored: analysis/average_cor_gtex_2_cache/ Ignored: analysis/average_cor_gtex_cache/ Ignored: analysis/brca_cache/ Ignored: analysis/cash_deconv_cache/ Ignored: analysis/cash_fdr_1_cache/ Ignored: analysis/cash_fdr_2_cache/ Ignored: analysis/cash_fdr_3_cache/ Ignored: analysis/cash_fdr_4_cache/ Ignored: analysis/cash_fdr_5_cache/ Ignored: analysis/cash_fdr_6_cache/ Ignored: analysis/cash_plots_cache/ Ignored: analysis/cash_sim_1_cache/ Ignored: analysis/cash_sim_2_cache/ Ignored: analysis/cash_sim_3_cache/ Ignored: analysis/cash_sim_4_cache/ Ignored: analysis/cash_sim_5_cache/ Ignored: analysis/cash_sim_6_cache/ Ignored: analysis/cash_sim_7_cache/ Ignored: analysis/correlated_z_2_cache/ Ignored: analysis/correlated_z_3_cache/ Ignored: analysis/correlated_z_cache/ Ignored: analysis/create_null_cache/ Ignored: analysis/cutoff_null_cache/ Ignored: analysis/design_matrix_2_cache/ Ignored: analysis/design_matrix_cache/ Ignored: analysis/diagnostic_ash_cache/ Ignored: analysis/diagnostic_correlated_z_2_cache/ Ignored: analysis/diagnostic_correlated_z_3_cache/ Ignored: analysis/diagnostic_correlated_z_cache/ Ignored: analysis/diagnostic_plot_2_cache/ Ignored: analysis/diagnostic_plot_cache/ Ignored: analysis/efron_leukemia_cache/ Ignored: analysis/fitting_normal_cache/ Ignored: analysis/gaussian_derivatives_2_cache/ Ignored: analysis/gaussian_derivatives_3_cache/ Ignored: analysis/gaussian_derivatives_4_cache/ Ignored: analysis/gaussian_derivatives_5_cache/ Ignored: analysis/gaussian_derivatives_cache/ Ignored: analysis/gd-ash_cache/ Ignored: analysis/gd_delta_cache/ Ignored: analysis/gd_lik_2_cache/ Ignored: analysis/gd_lik_cache/ Ignored: analysis/gd_w_cache/ Ignored: analysis/knockoff_10_cache/ Ignored: analysis/knockoff_2_cache/ Ignored: analysis/knockoff_3_cache/ Ignored: analysis/knockoff_4_cache/ Ignored: analysis/knockoff_5_cache/ Ignored: analysis/knockoff_6_cache/ Ignored: analysis/knockoff_7_cache/ Ignored: analysis/knockoff_8_cache/ Ignored: analysis/knockoff_9_cache/ Ignored: analysis/knockoff_cache/ Ignored: analysis/knockoff_var_cache/ Ignored: analysis/marginal_z_alternative_cache/ Ignored: analysis/marginal_z_cache/ Ignored: analysis/mosek_reg_2_cache/ Ignored: analysis/mosek_reg_4_cache/ Ignored: analysis/mosek_reg_5_cache/ Ignored: analysis/mosek_reg_6_cache/ Ignored: analysis/mosek_reg_cache/ Ignored: analysis/pihat0_null_cache/ Ignored: analysis/plot_diagnostic_cache/ Ignored: analysis/poster_obayes17_cache/ Ignored: analysis/real_data_simulation_2_cache/ Ignored: analysis/real_data_simulation_3_cache/ Ignored: analysis/real_data_simulation_4_cache/ Ignored: analysis/real_data_simulation_5_cache/ Ignored: analysis/real_data_simulation_cache/ Ignored: analysis/rmosek_primal_dual_2_cache/ Ignored: analysis/rmosek_primal_dual_cache/ Ignored: analysis/seqgendiff_cache/ Ignored: analysis/simulated_correlated_null_2_cache/ Ignored: analysis/simulated_correlated_null_3_cache/ Ignored: analysis/simulated_correlated_null_cache/ Ignored: analysis/simulation_real_se_2_cache/ Ignored: analysis/simulation_real_se_cache/ Ignored: analysis/smemo_2_cache/ Ignored: data/LSI/ Ignored: docs/.DS_Store Ignored: docs/figure/.DS_Store Ignored: output/fig/
Expand here to see past versions:
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | e05bc83 | LSun | 2018-05-12 | Update to 1.0 |
| rmd | cc0ab83 | Lei Sun | 2018-05-11 | update |
| html | 21e639d | LSun | 2018-04-05 | Build site. |
| rmd | 1187399 | LSun | 2018-04-05 | wflow_publish(“analysis/knockoff_var.rmd”) |
| rmd | 69aab5a | Lei Sun | 2018-04-03 | knockoff variability |
| html | 6af9b53 | LSun | 2018-04-03 | Build site. |
| rmd | 5bce84a | LSun | 2018-04-03 | wflow_publish(c(“knockoff_6.rmd”, “knockoff_var.rmd”)) |
Introduction
We hypothesize that Knockoff might have a potential problem: the variability of the actual FDP could be large, although the mean of FDP is theoretically and empirically controlled at FDR.
The reason is that Knockoff relies on the flip sign property of the test statistics \(W_j\) for all null variables. Actually the events \(\{W_j \geq t_j\}\) or \(\{W_j \leq -t_j\}\) are independent for all null variables. However, they are not identically distributed.
Therefore, if every null variable is “unique,” in the sense that its test statistic has a distinct distribution, for example, if it tends to be larger or in a different region than anyone else, then this test statistic could randomly be positive or negative, and affect the estimate of FDP for different random samples.
In common high-dimensional settings it’s usually not a big problem since in those simulations no null variables are unique, meaning that we would expect to see \(W_i\) and \(W_j\) showing up in regions symmetric at zero.
A simple illustration
\(X\) is \(30 \times 4\), \(X_1\) and \(X_3\) are “true” variables, and \(X_2\) and \(X_4\) are nulls. \(X_2\) is highly correlated with \(X_1\), \(\rho = 0.99\), whereas \(X_4\) is almost uncorrelated with \(X_3\), \(\rho = 0.01\).
Below is a plot of \(W_2\) vs \(W_4\). As we can see, \(|W_2|\) is usually much greater than \(|W_4|\), and in many samples, only \(W_2\) will show up in the rejection region \(W \geq t\) or its symmetric null estimation region \(W \le -t\), which brings variability of \(\hat{FDP}\) as the null statistic \(W_2\) shows up in one region or the other.

Example
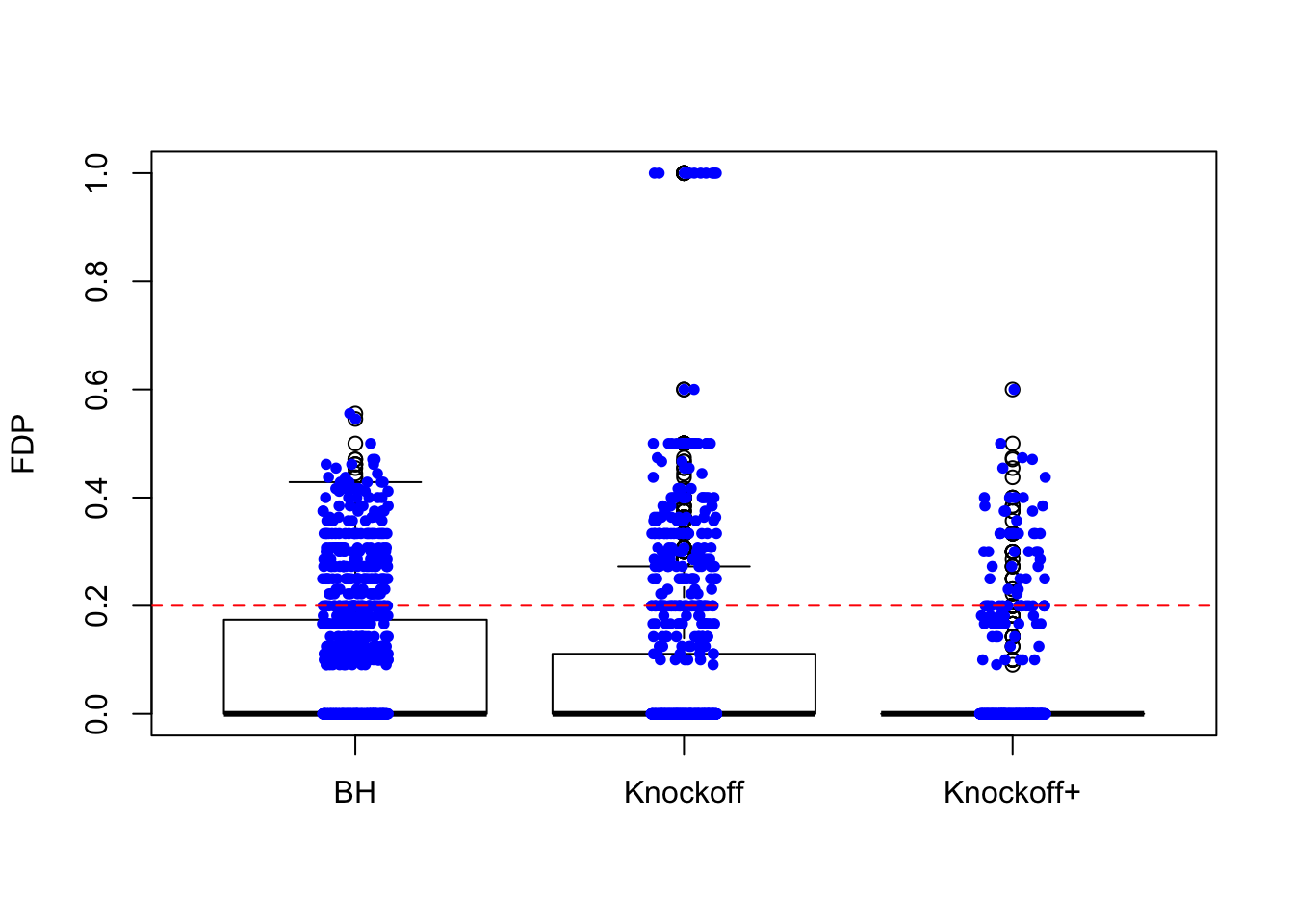
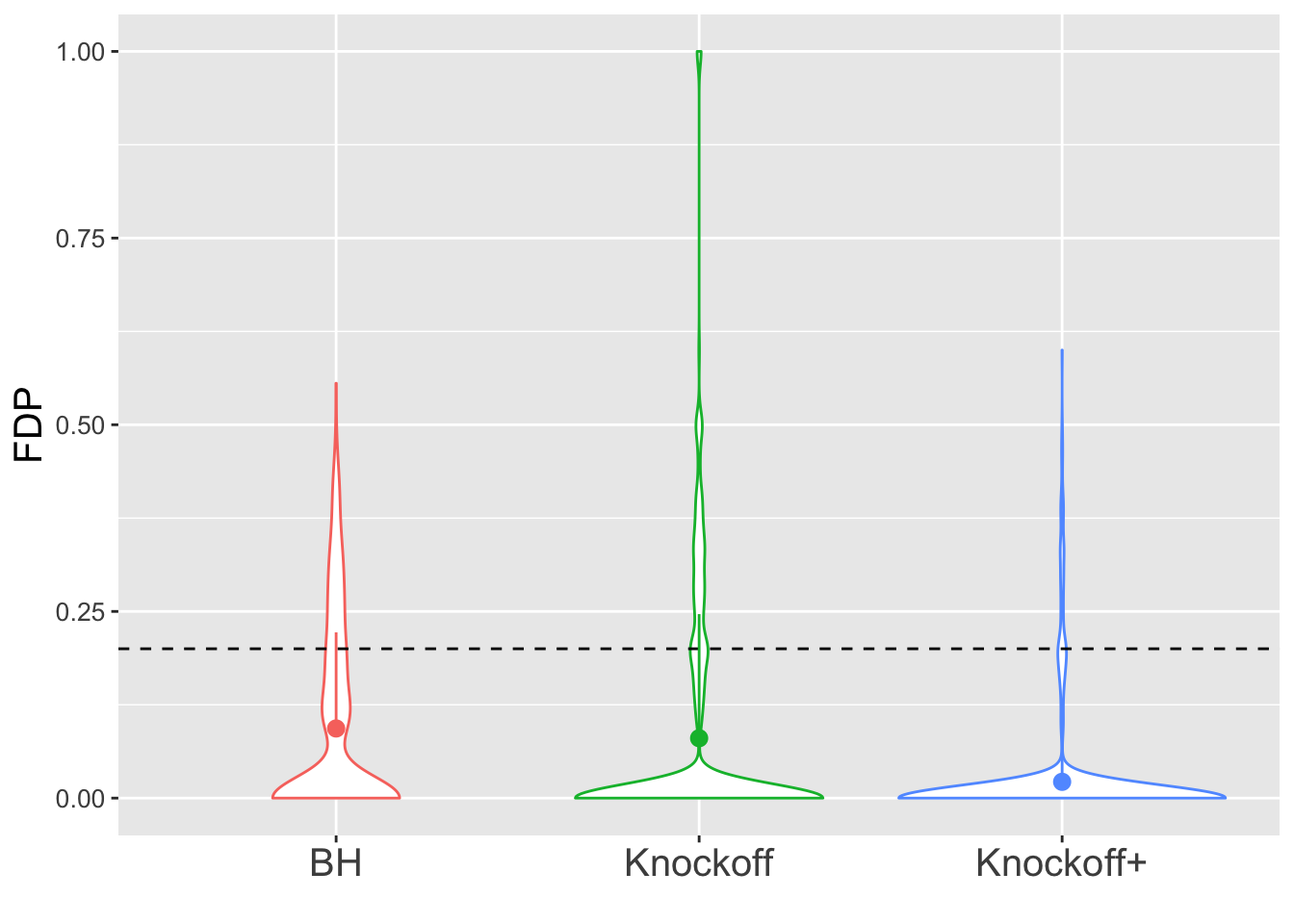
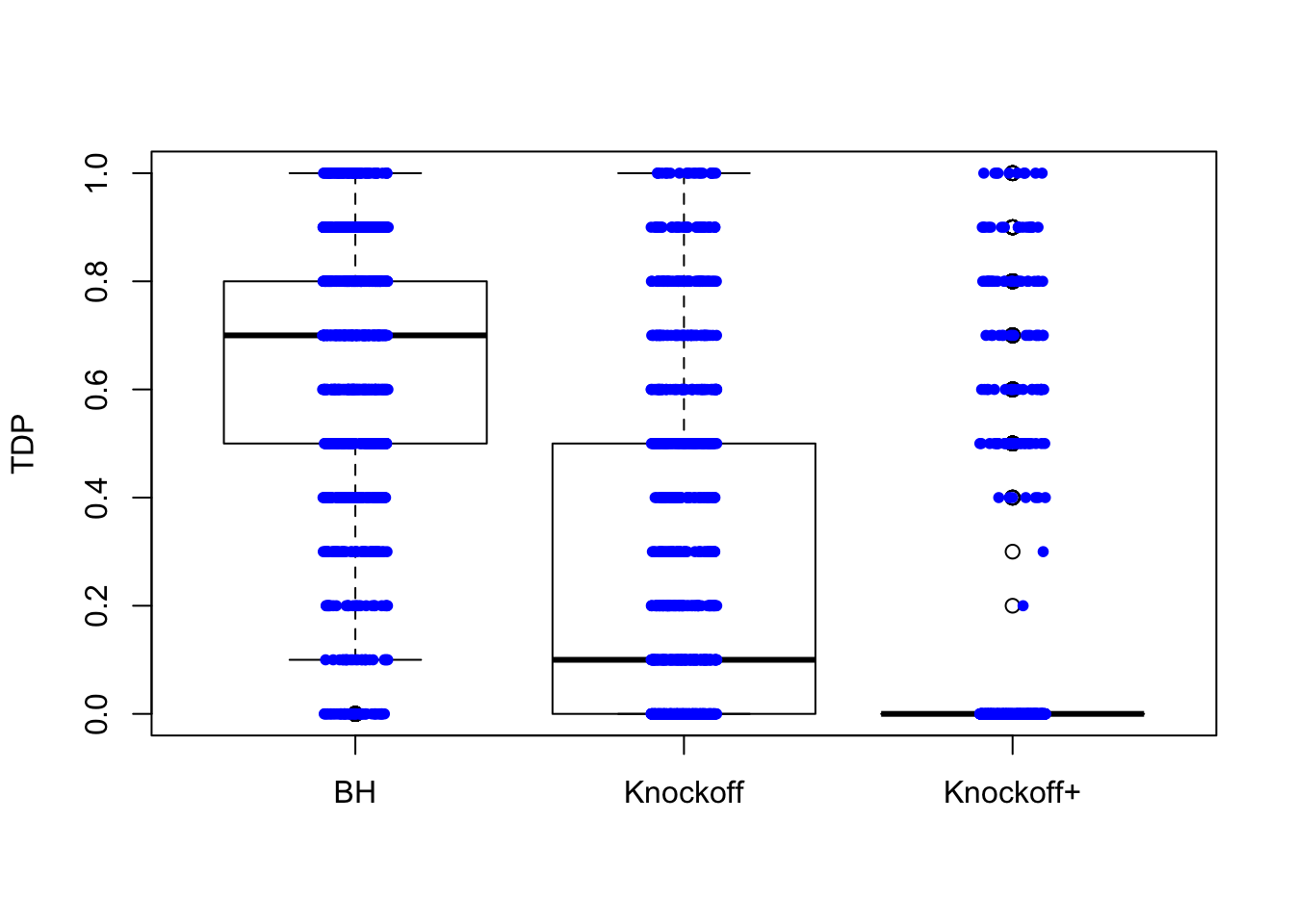
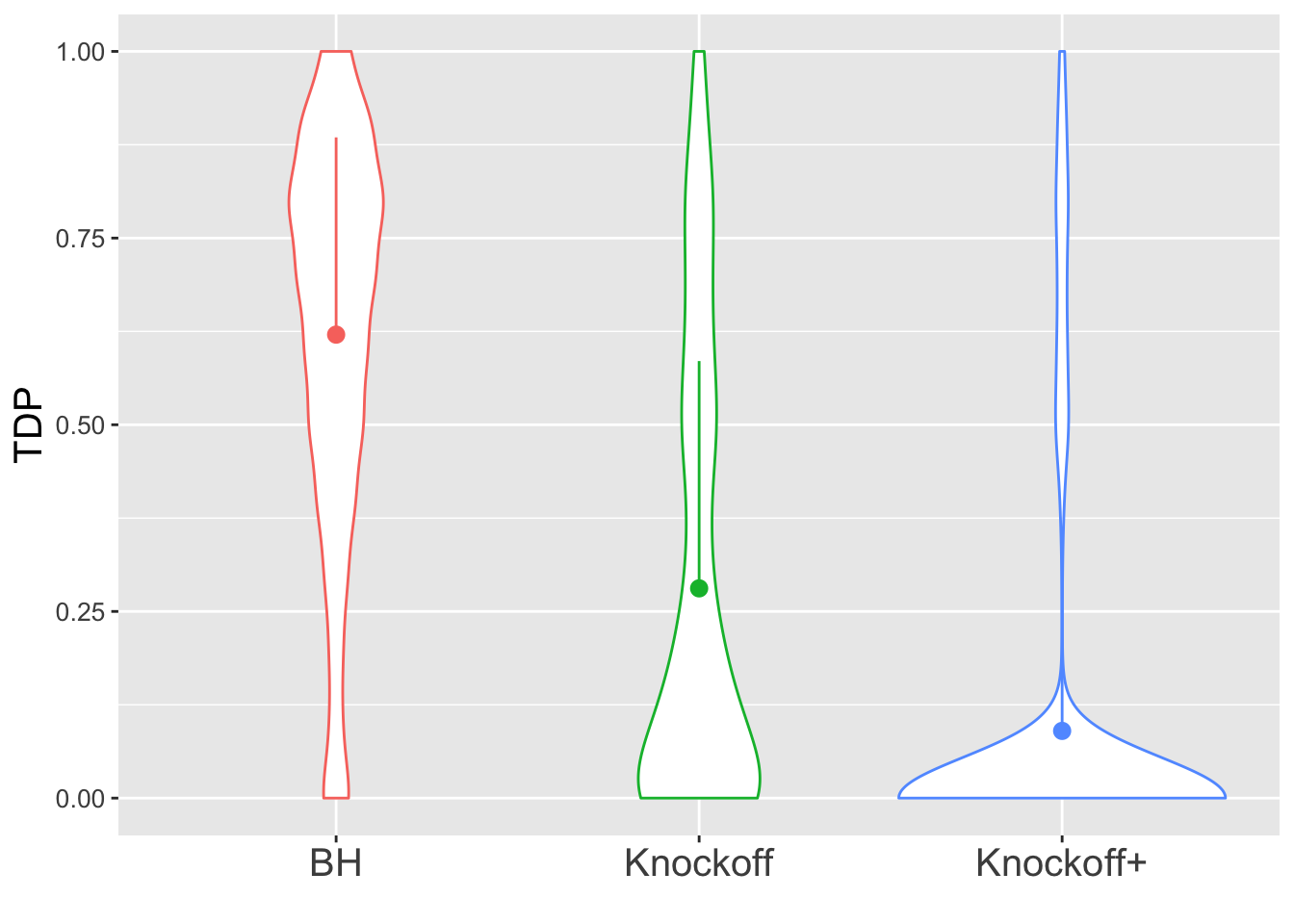
\(X\) is \(n \times 20\). There are \(10\) “true” variables: \(X_1, X_3, X_5, \ldots, X_{17}, X_{19}\) and \(10\) null variables: \(X_2, X_4, X_6, \ldots, X_{18}, X_{20}\). The correlation between the true and null variables are decreasing such that \(\rho(X_1, X_2) = 0.9, \rho(X_3, X_4) = 0.8, \rho(X_5, X_6) = 0.7, \ldots, \rho(X_{17}, X_{18}) = 0.2, \rho(X_{19}, X_{20}) = 0.1\), and all other pairwise correlations are zero.
\(n = 30\)
The variability in FDP given by Knockoff is obvious.
Expand here to see past versions of unnamed-chunk-4-1.png:
| Version | Author | Date |
|---|---|---|
| 21e639d | LSun | 2018-04-05 |
Expand here to see past versions of unnamed-chunk-4-2.png:
| Version | Author | Date |
|---|---|---|
| 21e639d | LSun | 2018-04-05 |
Expand here to see past versions of unnamed-chunk-4-3.png:
| Version | Author | Date |
|---|---|---|
| 21e639d | LSun | 2018-04-05 |
Expand here to see past versions of unnamed-chunk-4-4.png:
| Version | Author | Date |
|---|---|---|
| 21e639d | LSun | 2018-04-05 |
\(n = 10K\)
The variability in FDP given by Knockoff is less than when \(n = 30\), but still bigger than that given by BH.
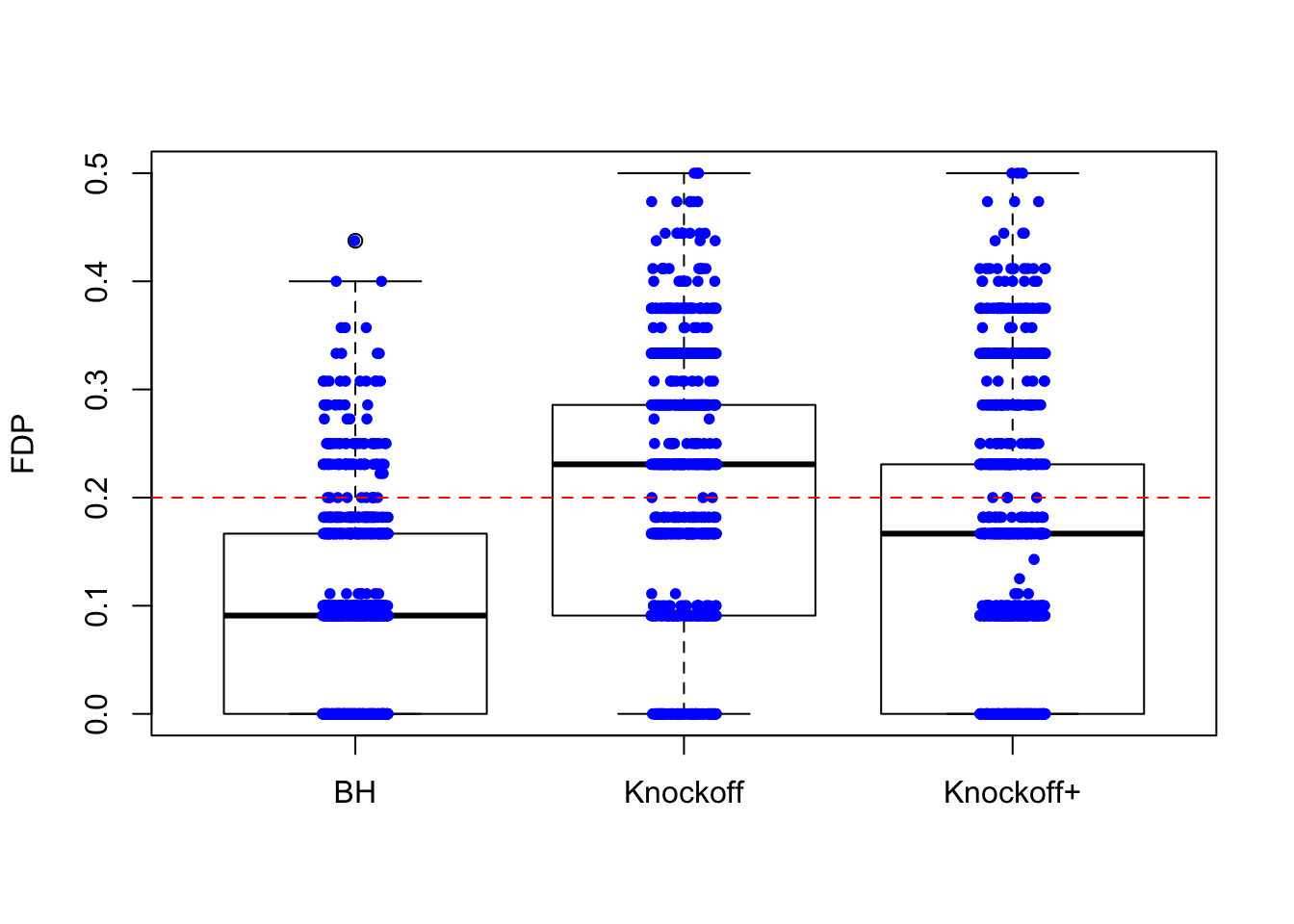
Expand here to see past versions of unnamed-chunk-5-1.png:
| Version | Author | Date |
|---|---|---|
| 21e639d | LSun | 2018-04-05 |
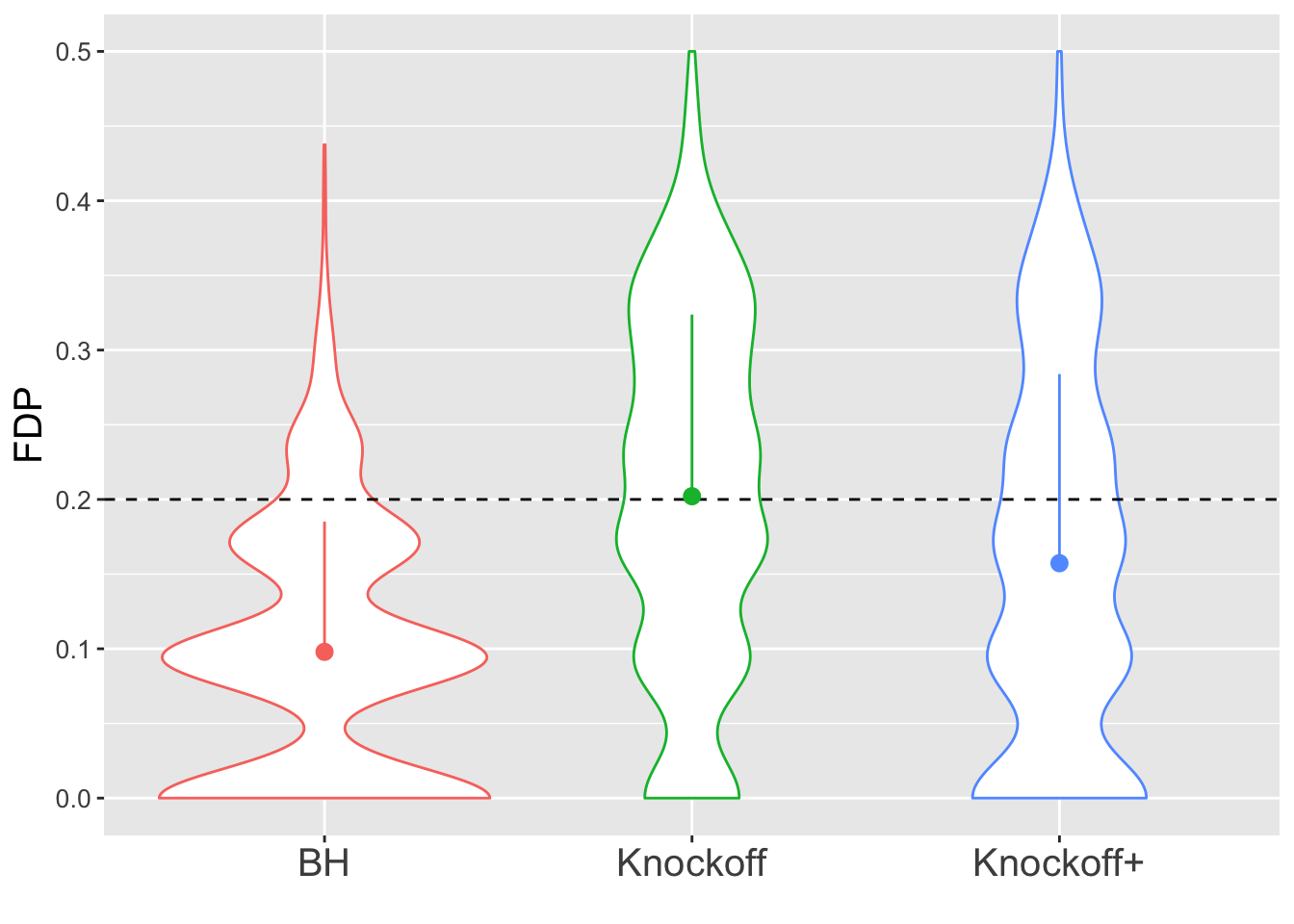
Expand here to see past versions of unnamed-chunk-5-2.png:
| Version | Author | Date |
|---|---|---|
| 21e639d | LSun | 2018-04-05 |
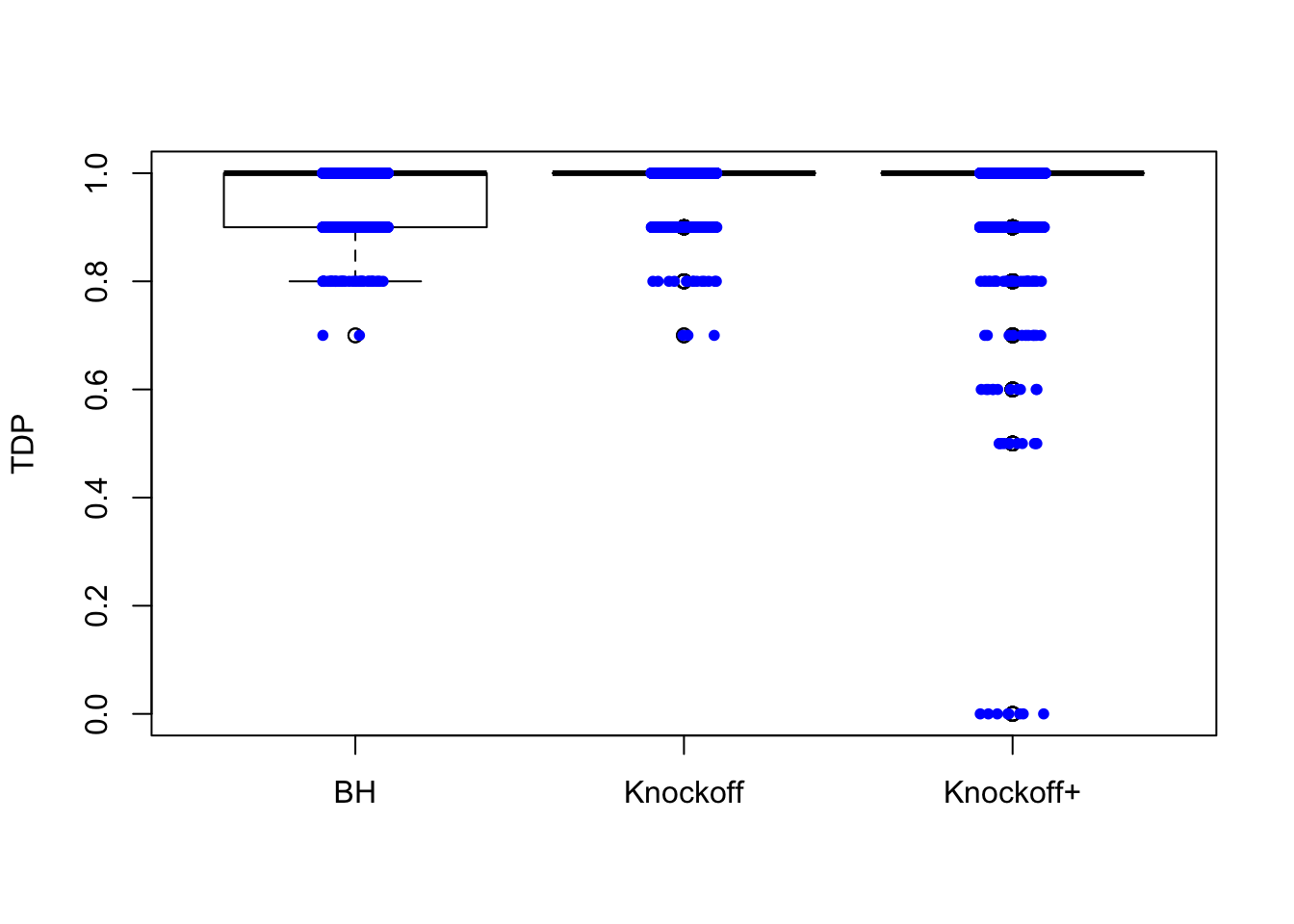
Expand here to see past versions of unnamed-chunk-5-3.png:
| Version | Author | Date |
|---|---|---|
| 21e639d | LSun | 2018-04-05 |
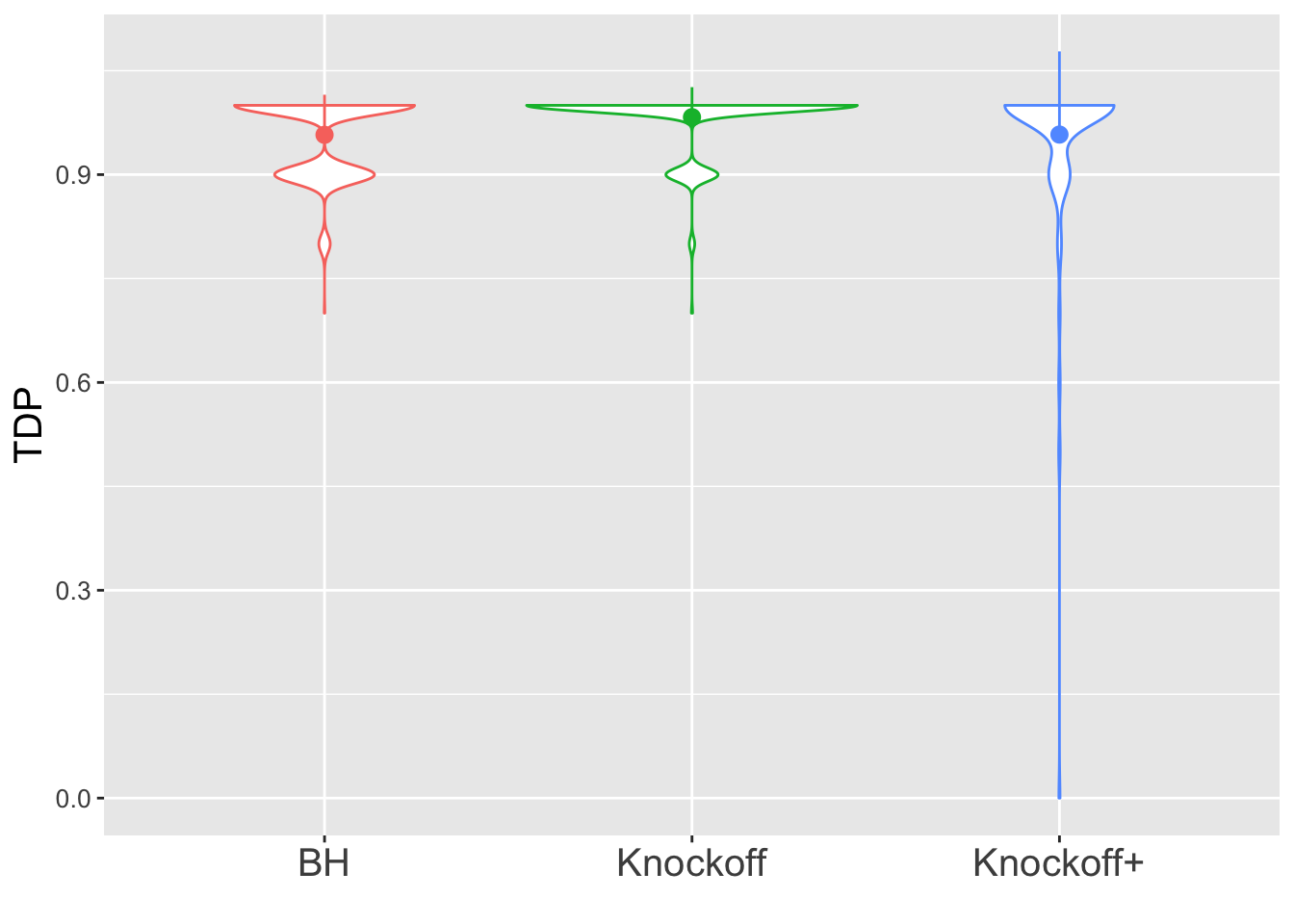
Expand here to see past versions of unnamed-chunk-5-4.png:
| Version | Author | Date |
|---|---|---|
| 21e639d | LSun | 2018-04-05 |
Session information
sessionInfo()R version 3.4.3 (2017-11-30)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS High Sierra 10.13.4
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] forcats_0.2.0 stringr_1.3.0 dplyr_0.7.4 purrr_0.2.4
[5] readr_1.1.1 tidyr_0.7.2 tibble_1.4.1 ggplot2_2.2.1
[9] tidyverse_1.2.1 Matrix_1.2-12 knockoff_0.3.0
loaded via a namespace (and not attached):
[1] reshape2_1.4.3 haven_1.1.0 lattice_0.20-35
[4] colorspace_1.3-2 htmltools_0.3.6 yaml_2.1.18
[7] rlang_0.1.6 R.oo_1.21.0 pillar_1.0.1
[10] foreign_0.8-69 glue_1.2.0 R.utils_2.6.0
[13] readxl_1.0.0 modelr_0.1.1 bindrcpp_0.2
[16] bindr_0.1 plyr_1.8.4 cellranger_1.1.0
[19] munsell_0.4.3 gtable_0.2.0 workflowr_1.0.1
[22] rvest_0.3.2 R.methodsS3_1.7.1 psych_1.7.8
[25] evaluate_0.10.1 labeling_0.3 knitr_1.20
[28] parallel_3.4.3 broom_0.4.3 Rcpp_0.12.16
[31] backports_1.1.2 scales_0.5.0 jsonlite_1.5
[34] mnormt_1.5-5 hms_0.4.0 digest_0.6.15
[37] stringi_1.1.6 grid_3.4.3 rprojroot_1.3-2
[40] cli_1.0.0 tools_3.4.3 magrittr_1.5
[43] lazyeval_0.2.1 crayon_1.3.4 whisker_0.3-2
[46] pkgconfig_2.0.1 xml2_1.1.1 lubridate_1.7.1
[49] rstudioapi_0.7 assertthat_0.2.0 rmarkdown_1.9
[52] httr_1.3.1 R6_2.2.2 nlme_3.1-131
[55] git2r_0.21.0 compiler_3.4.3 This reproducible R Markdown analysis was created with workflowr 1.0.1