Why AUC of BH and \(p\) values are different?
Lei Sun
2017-06-02
Last updated: 2018-05-12
workflowr checks: (Click a bullet for more information)-
✔ R Markdown file: up-to-date
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
-
✔ Environment: empty
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
-
✔ Seed:
set.seed(12345)The command
set.seed(12345)was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible. -
✔ Session information: recorded
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
-
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.✔ Repository version: 5b37742
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can usewflow_publishorwflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.Ignored files: Ignored: .DS_Store Ignored: .Rhistory Ignored: .Rproj.user/ Ignored: analysis/.DS_Store Ignored: analysis/BH_robustness_cache/ Ignored: analysis/FDR_Null_cache/ Ignored: analysis/FDR_null_betahat_cache/ Ignored: analysis/Rmosek_cache/ Ignored: analysis/StepDown_cache/ Ignored: analysis/alternative2_cache/ Ignored: analysis/alternative_cache/ Ignored: analysis/ash_gd_cache/ Ignored: analysis/average_cor_gtex_2_cache/ Ignored: analysis/average_cor_gtex_cache/ Ignored: analysis/brca_cache/ Ignored: analysis/cash_deconv_cache/ Ignored: analysis/cash_fdr_1_cache/ Ignored: analysis/cash_fdr_2_cache/ Ignored: analysis/cash_fdr_3_cache/ Ignored: analysis/cash_fdr_4_cache/ Ignored: analysis/cash_fdr_5_cache/ Ignored: analysis/cash_fdr_6_cache/ Ignored: analysis/cash_plots_cache/ Ignored: analysis/cash_plots_fdp_cache/ Ignored: analysis/cash_sim_1_cache/ Ignored: analysis/cash_sim_2_cache/ Ignored: analysis/cash_sim_3_cache/ Ignored: analysis/cash_sim_4_cache/ Ignored: analysis/cash_sim_5_cache/ Ignored: analysis/cash_sim_6_cache/ Ignored: analysis/cash_sim_7_cache/ Ignored: analysis/correlated_z_2_cache/ Ignored: analysis/correlated_z_3_cache/ Ignored: analysis/correlated_z_cache/ Ignored: analysis/create_null_cache/ Ignored: analysis/cutoff_null_cache/ Ignored: analysis/design_matrix_2_cache/ Ignored: analysis/design_matrix_cache/ Ignored: analysis/diagnostic_ash_cache/ Ignored: analysis/diagnostic_correlated_z_2_cache/ Ignored: analysis/diagnostic_correlated_z_3_cache/ Ignored: analysis/diagnostic_correlated_z_cache/ Ignored: analysis/diagnostic_plot_2_cache/ Ignored: analysis/diagnostic_plot_cache/ Ignored: analysis/efron_leukemia_cache/ Ignored: analysis/fitting_normal_cache/ Ignored: analysis/gaussian_derivatives_2_cache/ Ignored: analysis/gaussian_derivatives_3_cache/ Ignored: analysis/gaussian_derivatives_4_cache/ Ignored: analysis/gaussian_derivatives_5_cache/ Ignored: analysis/gaussian_derivatives_cache/ Ignored: analysis/gd-ash_cache/ Ignored: analysis/gd_delta_cache/ Ignored: analysis/gd_lik_2_cache/ Ignored: analysis/gd_lik_cache/ Ignored: analysis/gd_w_cache/ Ignored: analysis/knockoff_10_cache/ Ignored: analysis/knockoff_2_cache/ Ignored: analysis/knockoff_3_cache/ Ignored: analysis/knockoff_4_cache/ Ignored: analysis/knockoff_5_cache/ Ignored: analysis/knockoff_6_cache/ Ignored: analysis/knockoff_7_cache/ Ignored: analysis/knockoff_8_cache/ Ignored: analysis/knockoff_9_cache/ Ignored: analysis/knockoff_cache/ Ignored: analysis/knockoff_var_cache/ Ignored: analysis/marginal_z_alternative_cache/ Ignored: analysis/marginal_z_cache/ Ignored: analysis/mosek_reg_2_cache/ Ignored: analysis/mosek_reg_4_cache/ Ignored: analysis/mosek_reg_5_cache/ Ignored: analysis/mosek_reg_6_cache/ Ignored: analysis/mosek_reg_cache/ Ignored: analysis/pihat0_null_cache/ Ignored: analysis/plot_diagnostic_cache/ Ignored: analysis/poster_obayes17_cache/ Ignored: analysis/real_data_simulation_2_cache/ Ignored: analysis/real_data_simulation_3_cache/ Ignored: analysis/real_data_simulation_4_cache/ Ignored: analysis/real_data_simulation_5_cache/ Ignored: analysis/real_data_simulation_cache/ Ignored: analysis/rmosek_primal_dual_2_cache/ Ignored: analysis/rmosek_primal_dual_cache/ Ignored: analysis/seqgendiff_cache/ Ignored: analysis/simulated_correlated_null_2_cache/ Ignored: analysis/simulated_correlated_null_3_cache/ Ignored: analysis/simulated_correlated_null_cache/ Ignored: analysis/simulation_real_se_2_cache/ Ignored: analysis/simulation_real_se_cache/ Ignored: analysis/smemo_2_cache/ Ignored: data/LSI/ Ignored: docs/.DS_Store Ignored: docs/figure/.DS_Store Ignored: output/fig/ Untracked files: Untracked: analysis/cash_plots_fdp_files/ Untracked: docs/cash_plots_fdp_files/
Expand here to see past versions:
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| rmd | cc0ab83 | Lei Sun | 2018-05-11 | update |
| html | 0f36d99 | LSun | 2017-12-21 | Build site. |
| html | 853a484 | LSun | 2017-11-07 | Build site. |
| html | 82f2a7d | LSun | 2017-06-02 | auc |
| rmd | bf20fcd | LSun | 2017-06-02 | auc |
| rmd | a930ad9 | LSun | 2017-06-02 | BH |
| html | 7e2860d | LSun | 2017-06-02 | auc |
| rmd | 549ca68 | LSun | 2017-06-02 | auc |
Introduction
We usually use AUC (area under the receiver operating characteristic curve) to measure the performance of a statistical testing procedure. In essence, AUC is solely determined by how the hypotheses are ranked by the procedure. Thus, Matthew’s intuition is that the three methods based on \(p\) values, \(p\) values, Benjamini-Hochberg, and qvalue should give the equivalent AUC. However, it doesn’t appear so in our simulation. It turns out there is a simple explanation for the difference bwteen BH and \(p\) values.
A simple illustration
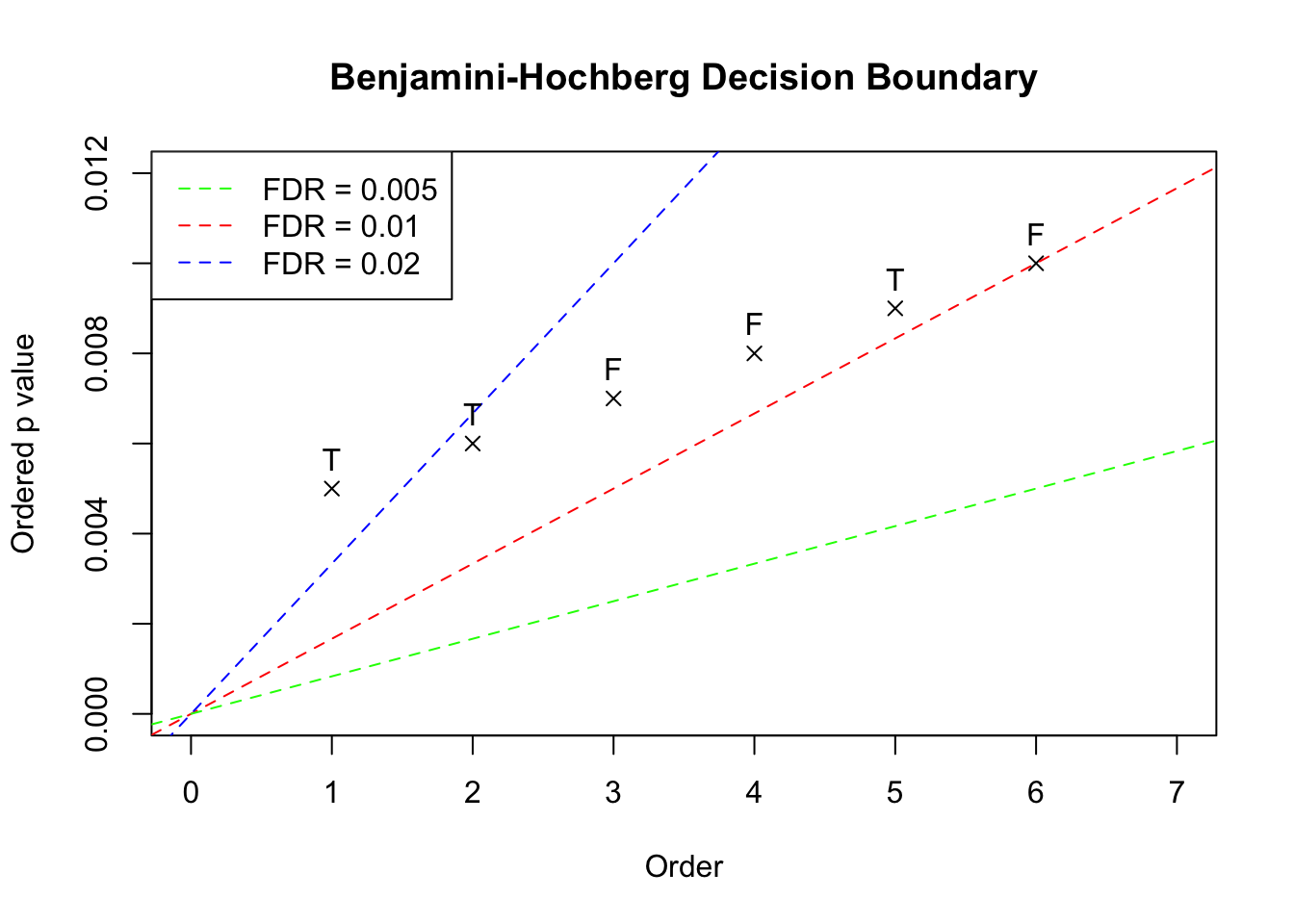
Given a nominal FDR level \(\alpha\), and \(m\) \(p\) values, BH starts with the least significant or largest \(p\) value and compares each ordered \(p\) value \(p_{(k)}\) with \(\frac{\alpha}mk\). Now suppose we have 6 \(p\) values in the increasing order, \(\left\{0.005, 0.006, 0.007, 0.008, 0.009, 0.010\right\}\), plotted as follows. These \(p\) values correpond with hypotheses of true positives (“T”) or false positives (“F”). In this example, the hypotheses associated with the ordered \(p\) values are set to be \(\left\{T, T, F, F, T, F\right\}\).
Expand here to see past versions of unnamed-chunk-1-1.png:
| Version | Author | Date |
|---|---|---|
| 0f36d99 | LSun | 2017-12-21 |
| 7e2860d | LSun | 2017-06-02 |
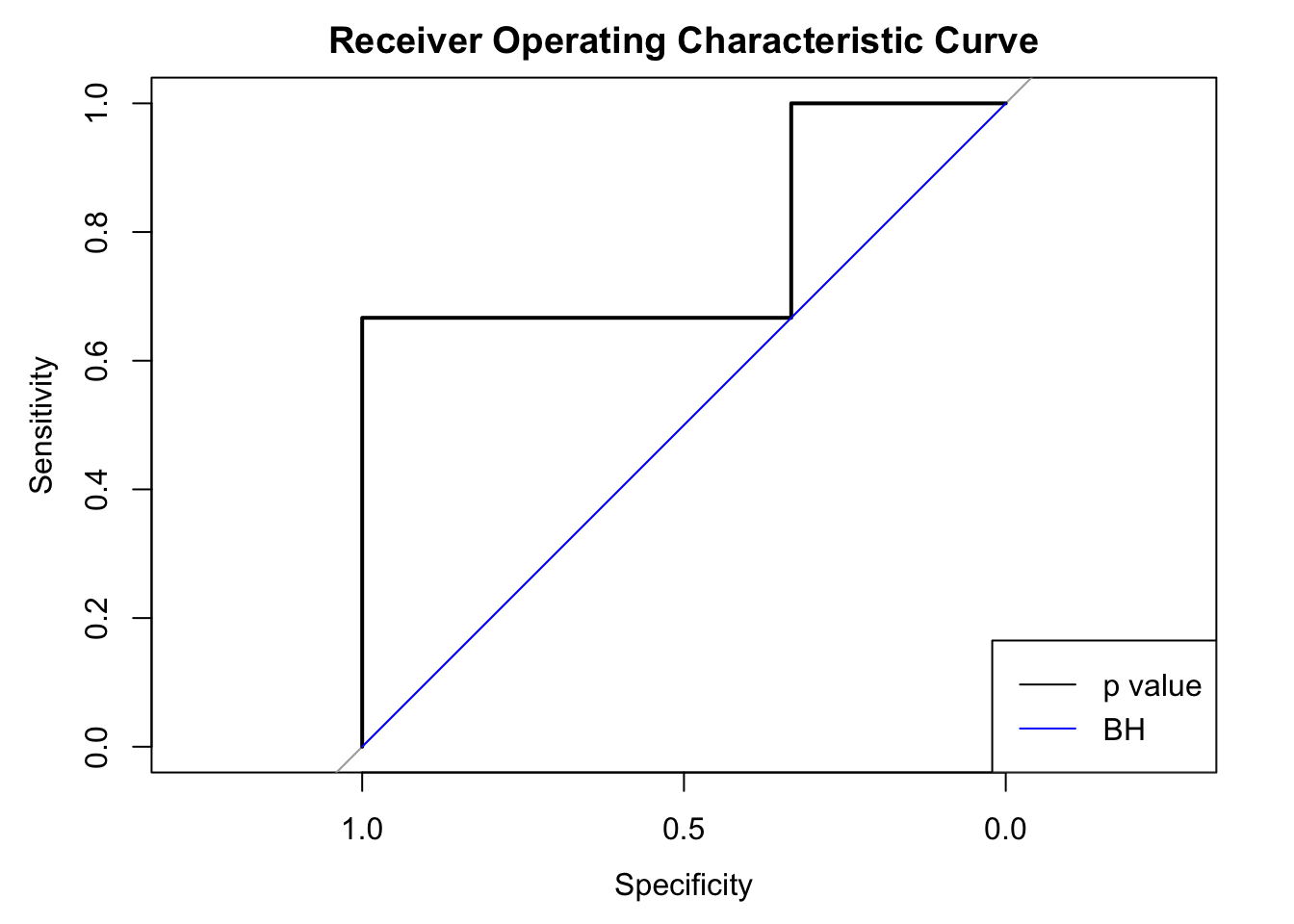
The three dotted lines in the plot show the three scenarios with BH applied to this data set. The critical decision boundary is when we have the nominal FDR \(\alpha = 0.01\) in red. When \(\alpha < 0.01\), like the green line, no \(p\) value falls below the line and thus no discovery is declared. On the other hand, when \(\alpha > 0.01\), like the blue line, the largest \(p\) value falls below the line and thus all hypotheses would be declared as discoveries. Therefore, under BH, at different nominal FDR levels, we either fail to reject any null hypothesis, or reject all of \(6\) together. In another word, we either have false positive rate (FPR) \(=\) true positive rate (TPR) \(=\) 0, or FRP \(=\) TPR \(=\) 1. Below we plot the two ROC curves by BH and by simply thresholding \(p\) values. In this case using \(p\) values (AUC \(=\) 0.78) is actually better than using BH (AUC = 0.5).
Expand here to see past versions of unnamed-chunk-2-1.png:
| Version | Author | Date |
|---|---|---|
| 0f36d99 | LSun | 2017-12-21 |
| 7e2860d | LSun | 2017-06-02 |
- Note that
BHindeed doesn’t change the order of hypotheses set by \(p\) values. As in this exmaple, if a hypothesis with a larger \(p\) value is rejected, all hypotheses with smaller \(p\) values will also be rejected; meanwhile, when a hypothesis with a smaller \(p\) value is not rejected, all hypotheses with larger \(p\) values won’t be rejected either. - However,
BHis capable of generating ties in hypotheses. As in this exmaple, all \(6\) hypothese have distinctive \(p\) values, yet they are essentially equivalent underBH.
This simple illustration shows why we would have different AUC with \(p\) values and BH.
Session information
sessionInfo()R version 3.4.3 (2017-11-30)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS High Sierra 10.13.4
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] pROC_1.10.0
loaded via a namespace (and not attached):
[1] Rcpp_0.12.16 knitr_1.20 whisker_0.3-2
[4] magrittr_1.5 workflowr_1.0.1 munsell_0.4.3
[7] colorspace_1.3-2 rlang_0.1.6 stringr_1.3.0
[10] plyr_1.8.4 tools_3.4.3 grid_3.4.3
[13] gtable_0.2.0 R.oo_1.21.0 git2r_0.21.0
[16] htmltools_0.3.6 lazyeval_0.2.1 yaml_2.1.18
[19] rprojroot_1.3-2 digest_0.6.15 tibble_1.4.1
[22] ggplot2_2.2.1 R.utils_2.6.0 evaluate_0.10.1
[25] rmarkdown_1.9 stringi_1.1.6 pillar_1.0.1
[28] compiler_3.4.3 scales_0.5.0 backports_1.1.2
[31] R.methodsS3_1.7.1
This reproducible R Markdown analysis was created with workflowr 1.0.1