\(\hat\pi_0\) on correlated global null estimated by ashr and truncash
Lei Sun
2017-02-21
Last updated: 2017-02-22
Code version: 242b122
Introduction
Using correlated global null data simulated from GTEx/Liver, \(\hat\pi_0\) is estimated by truncash and ashr. Ideally the estimates should be close to \(1\).
Simulation
In truncash, the threshold is \(T = 1.96\).
library(ashr)
source("../code/truncash.R")p = read.table("../output/p_null_liver_777.txt")
z = read.table("../output/z_null_liver_777.txt")
betahat = read.table("../output/betahat_null_liver_777.txt")
m = dim(p)[1]
n = dim(p)[2]
pihat0.ash = pihat0.truncash = c()
for (i in 1:m) {
betahat_trial = as.numeric(betahat[i, ])
sebetahat_trial = - betahat_trial / as.numeric(z[i, ])
fit.ash = ashr::ash(betahat_trial, sebetahat_trial, method = "fdr", mixcompdist = "normal")
pihat0.ash[i] = get_pi0(fit.ash)
fit.truncash = truncash(betahat_trial, sebetahat_trial, t = qnorm(0.975))
pihat0.truncash[i] = get_pi0(fit.truncash)
}Result
False positive rate by ash
mean(pihat0.ash < 1)[1] 0.287False positive rate by truncash
mean(pihat0.truncash < 1)[1] 0.202Compare \(\hat\pi_0\) estimated by ash and truncash
xlim = c(min(c(pihat0.ash, pihat0.truncash)), 1)
plot(pihat0.ash, pihat0.truncash, xlim = xlim, ylim = xlim, xlab = "ash", ylab = "truncash", main = expression(hat(pi)[0]))
abline(0, 1, lty = 2, col = "blue")
abline(v = 1, lty = 3, col = "red")
abline(h = 1, lty = 3, col = "red")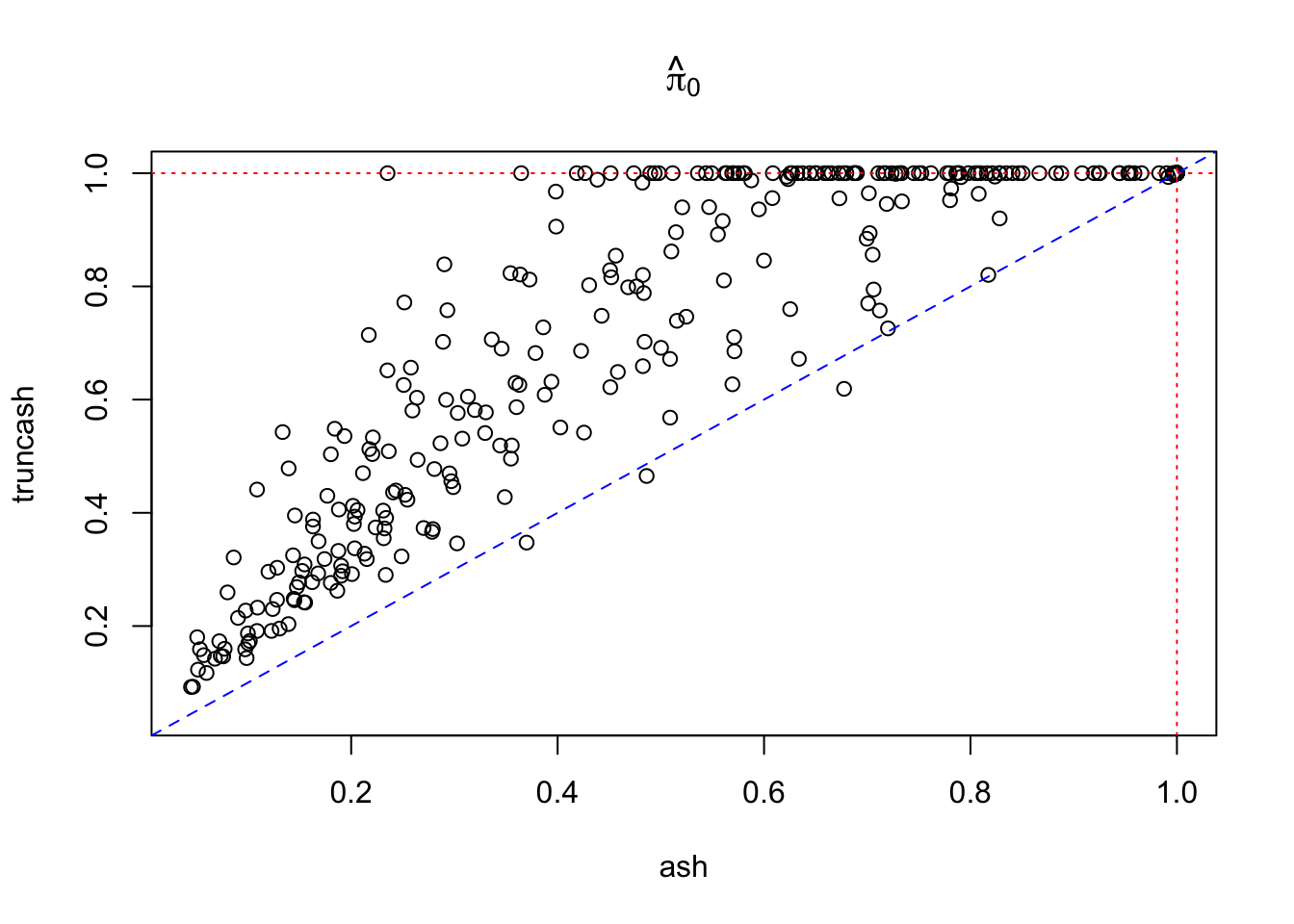
boxplot(pihat0.ash, pihat0.truncash, main = expression(hat(pi)[0]), names = c("ash", "truncash"))
abline(h = 1, lty = 2, col = "red")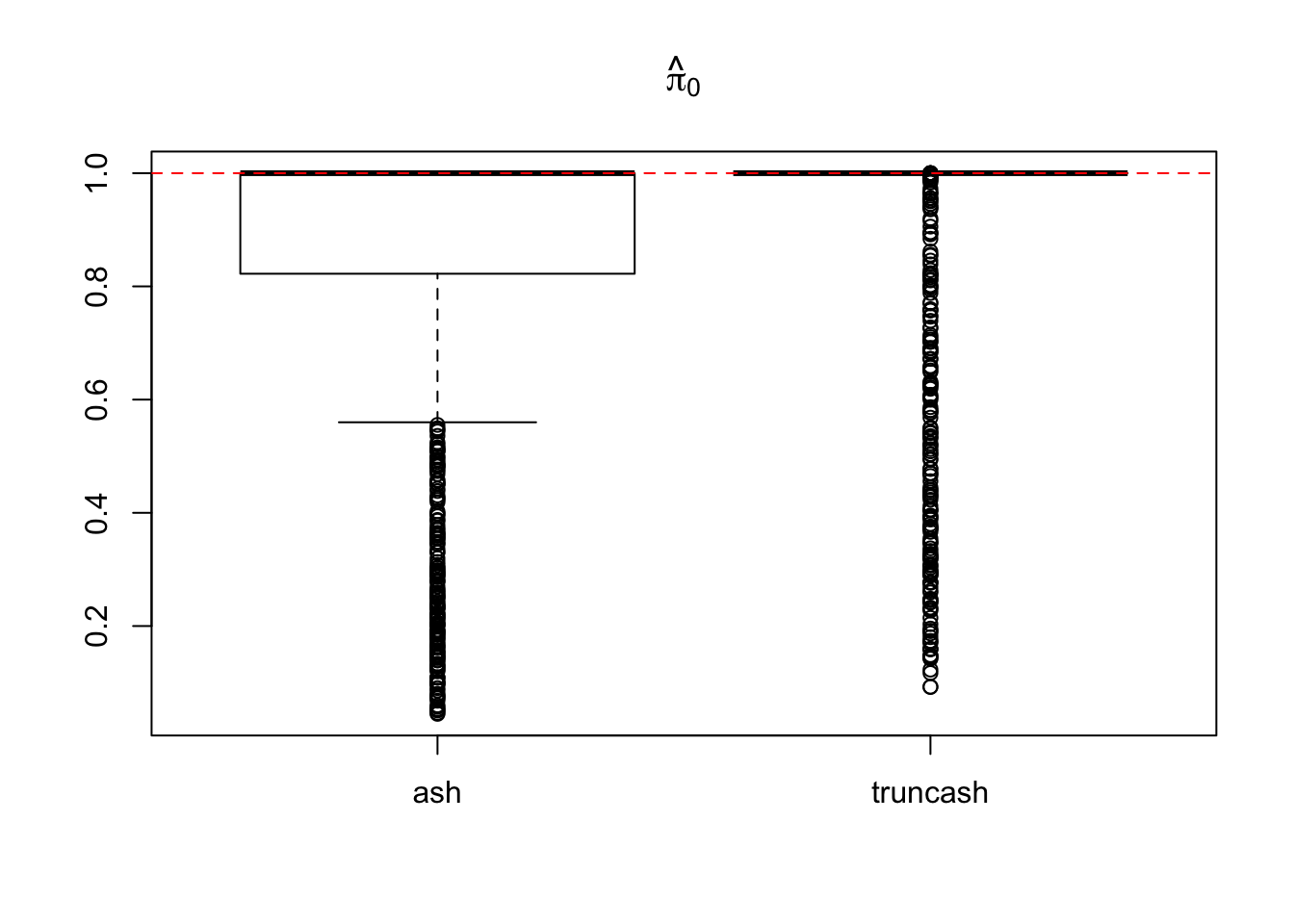
Session Information
sessionInfo()R version 3.3.2 (2016-10-31)
Platform: x86_64-apple-darwin13.4.0 (64-bit)
Running under: macOS Sierra 10.12.3
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] SQUAREM_2016.10-1 ashr_2.1.2
loaded via a namespace (and not attached):
[1] Rcpp_0.12.9 knitr_1.15.1 magrittr_1.5
[4] workflowr_0.3.0 MASS_7.3-45 doParallel_1.0.10
[7] pscl_1.4.9 lattice_0.20-34 foreach_1.4.3
[10] stringr_1.1.0 tools_3.3.2 parallel_3.3.2
[13] grid_3.3.2 git2r_0.18.0 htmltools_0.3.5
[16] iterators_1.0.8 yaml_2.1.14 rprojroot_1.2
[19] digest_0.6.9 codetools_0.2-15 evaluate_0.10
[22] rmarkdown_1.3 stringi_1.1.2 backports_1.0.5
[25] truncnorm_1.0-7 This R Markdown site was created with workflowr