Knockoff vs Bayesian: Large signals
Lei Sun
2018-04-10
Last updated: 2018-04-16
Code version: c7c5984
Introduction
varbvs.get.lfsr <- function (fit) {
# For each variable, and each hyperparameter setting, get the
# posterior probability that the regression coefficient is exactly
# zero.
p0 <- 1 - fit$alpha
# For each variable, and for each hyperparameter setting, get the
# posterior probability that the regression coefficient is negative.
pn <- with(fit,alpha * pnorm(0,mu,sqrt(s)))
# For each variable, and for each hyperparameter setting, ompute the
# local false sign rate (LFSR) following the formula given in
# Matthew's Biostatistics paper, "False discovery rates: a new deal".
p <- nrow(fit$alpha)
k <- ncol(fit$alpha)
lfsr <- matrix(0,p,k)
b <- pn > 0.5*(1 - p0)
lfsr[b] <- 1 - pn[b]
lfsr[!b] <- p0[!b] + pn[!b]
# Average the average LFSR over the hyperparameter settings, weighted
# by the probability of each hyperparameter setting.
lfsr <- c(lfsr %*% fit$w)
return(lfsr)
}\(n > p\)
n <- 1200
p <- 1000
k <- 50
m <- 100
q <- 0.1Independent design
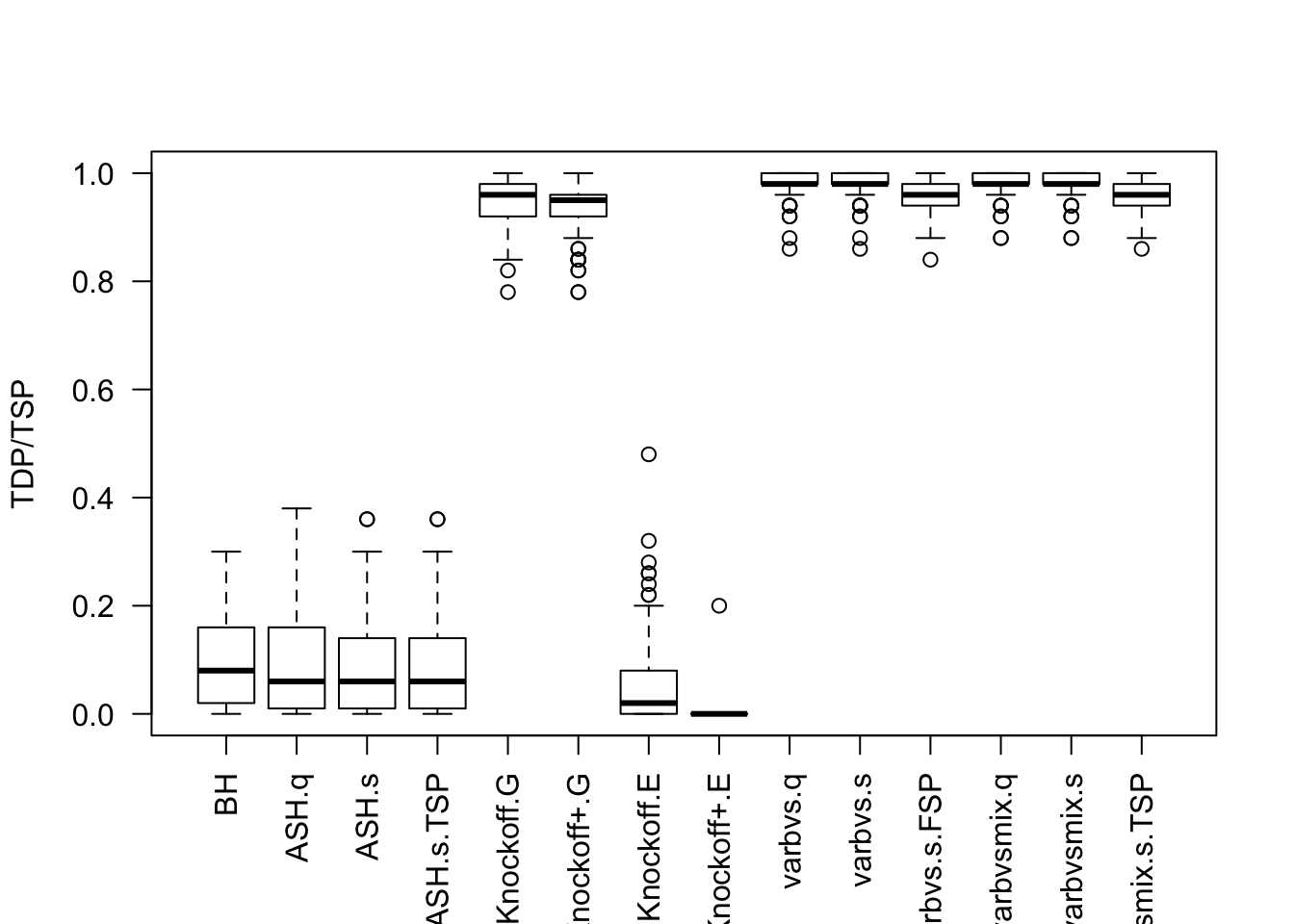
\(X_{n \times p}\) has independent columns simulated from \(N(0, (1/\sqrt n)^2)\) so they are roughly normalized.
Cov.X <- diag(1 / n, p)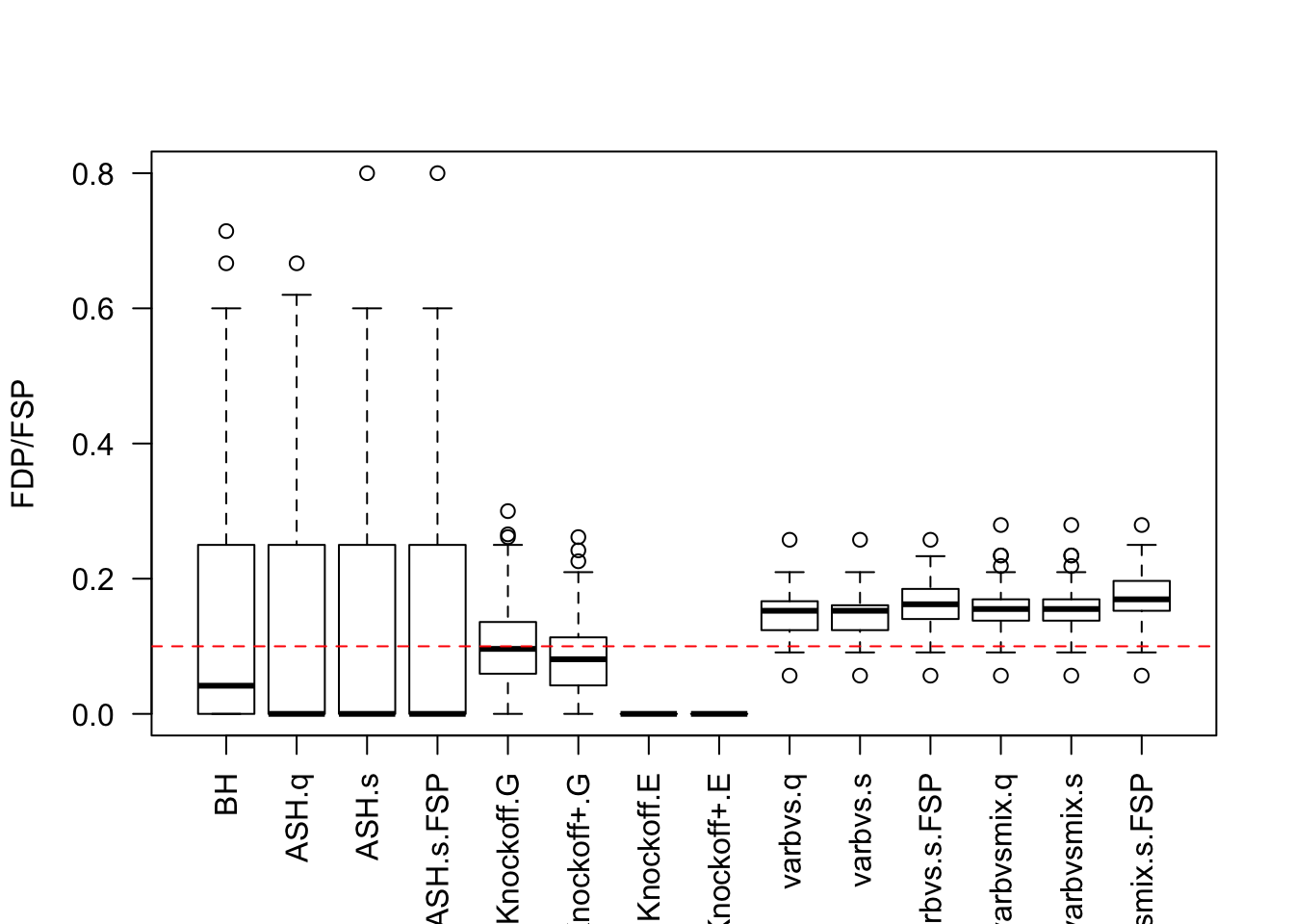
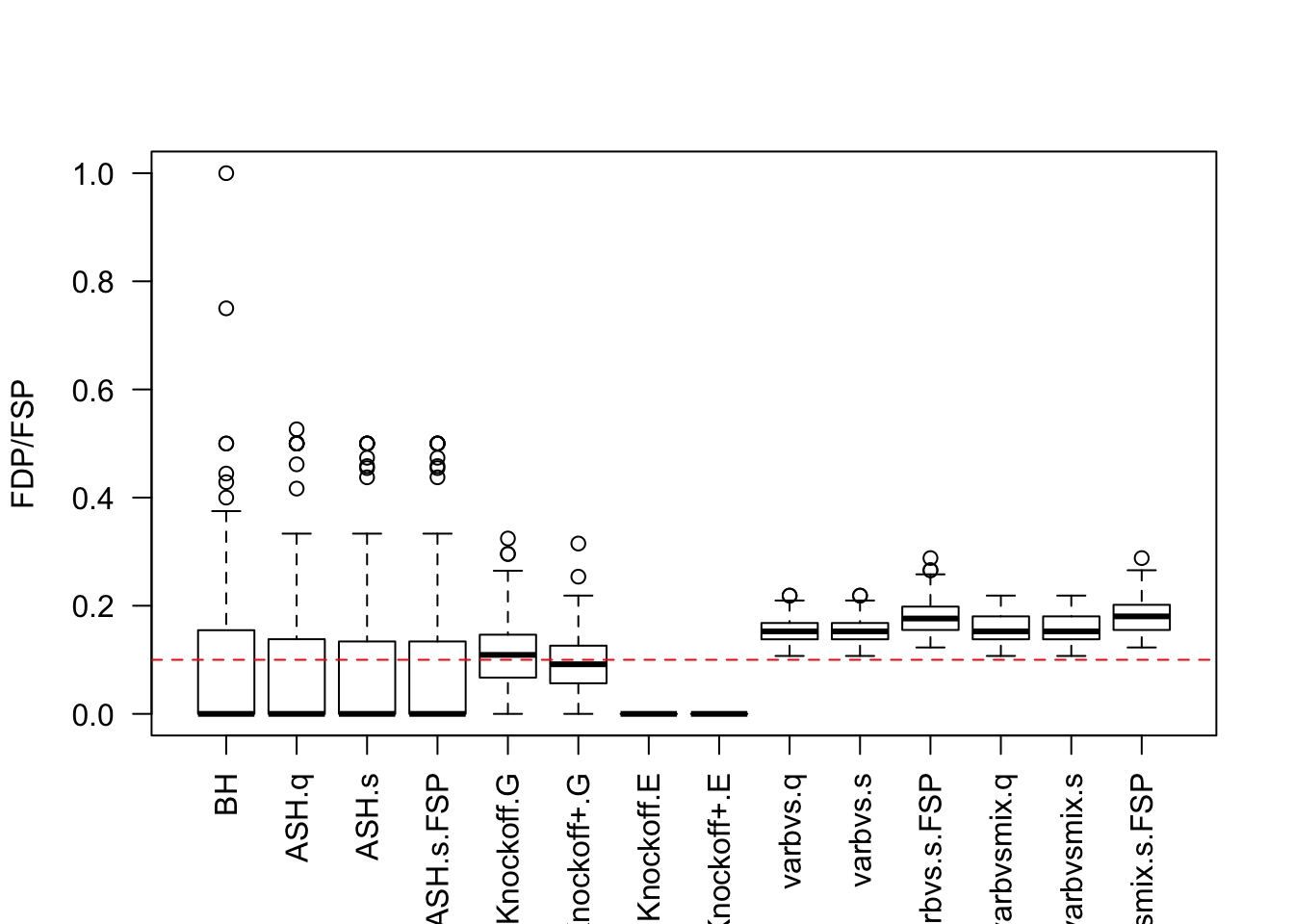
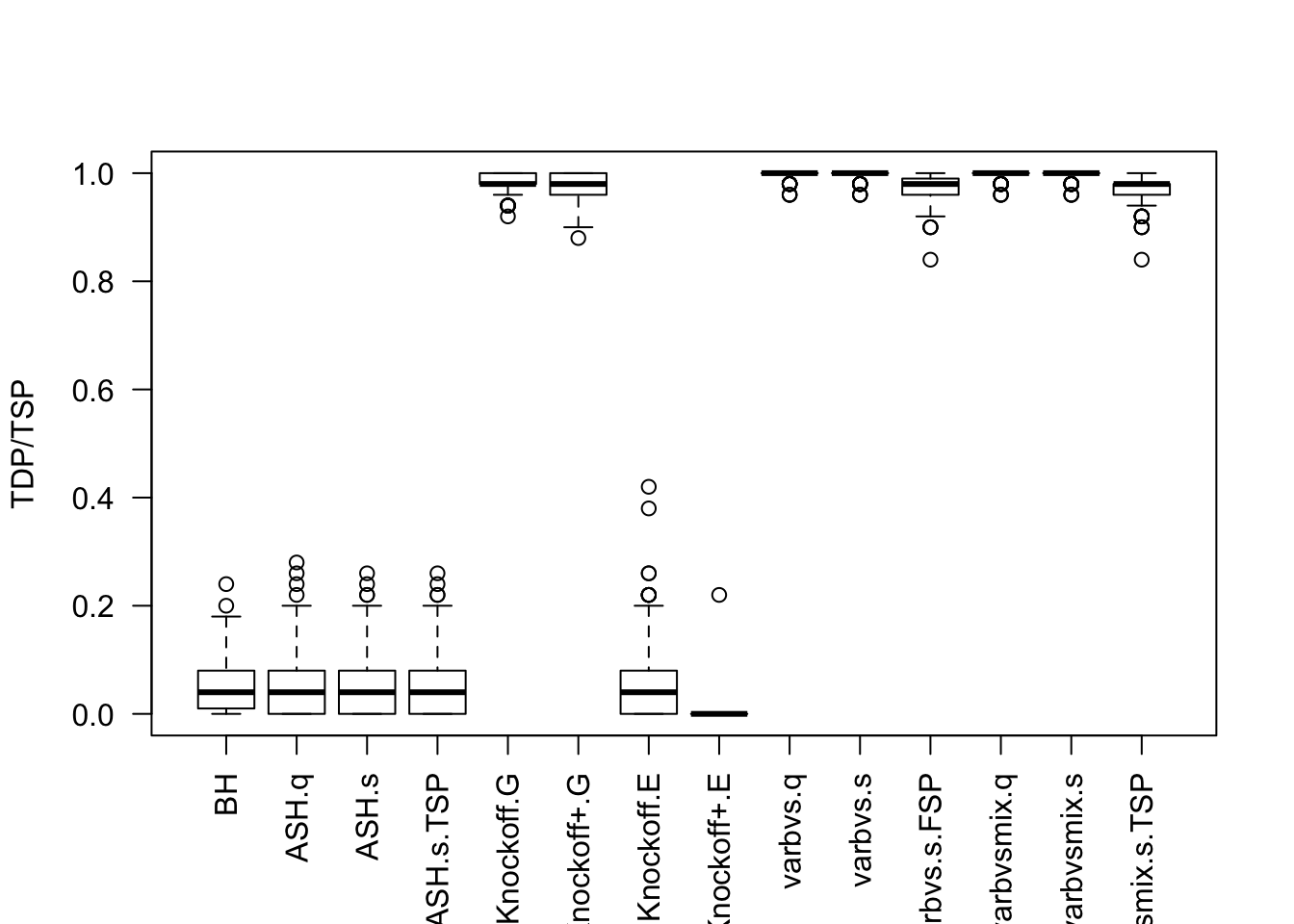
Local correlation / AR model for \(X\)
\(X_{n \times p}\) has correlation \(\Sigma_{ij} = \rho^{|i - j|}\). Each row is independently \(N(0, \frac1n\Sigma)\).
Factor Model for \(X\)
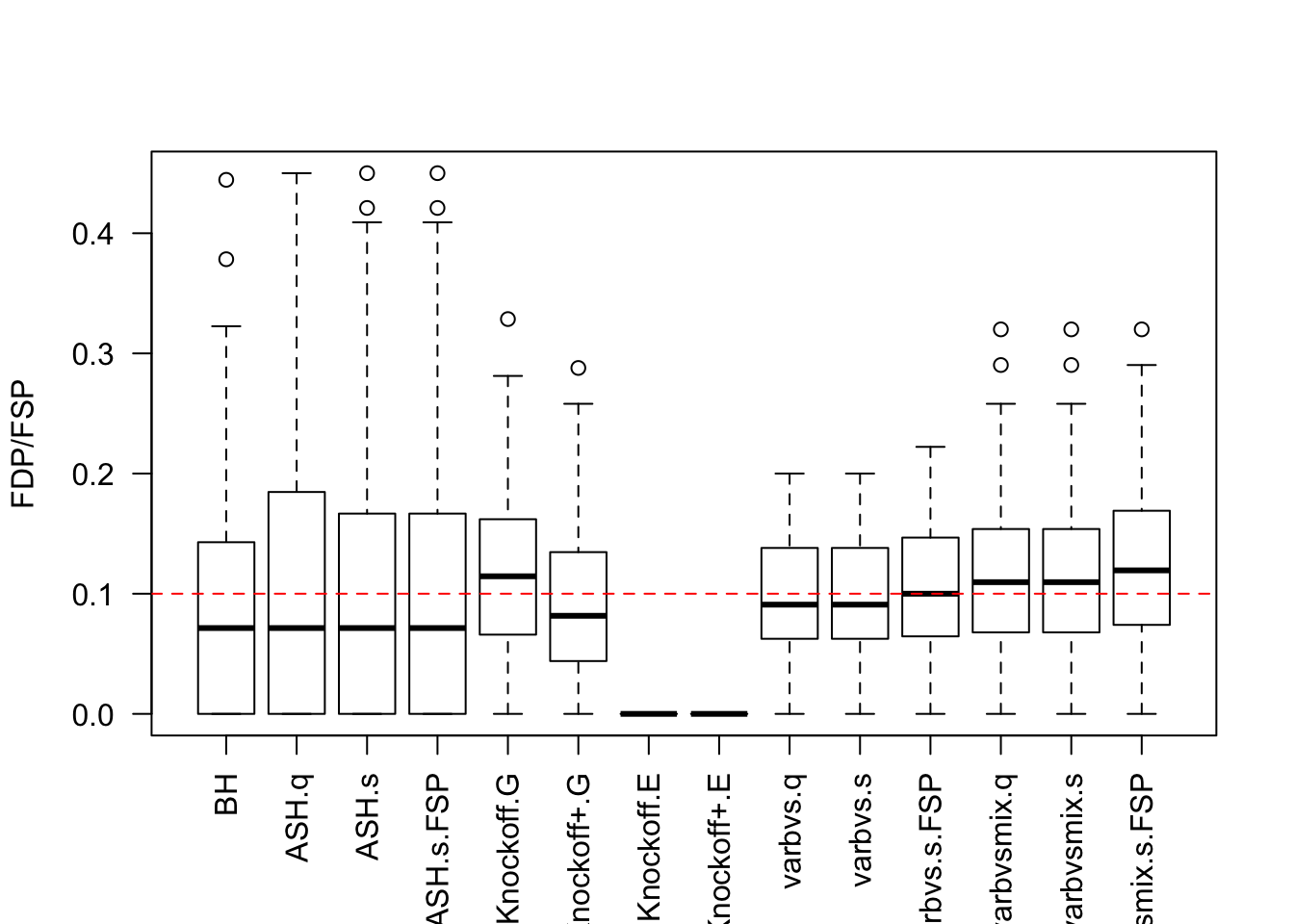
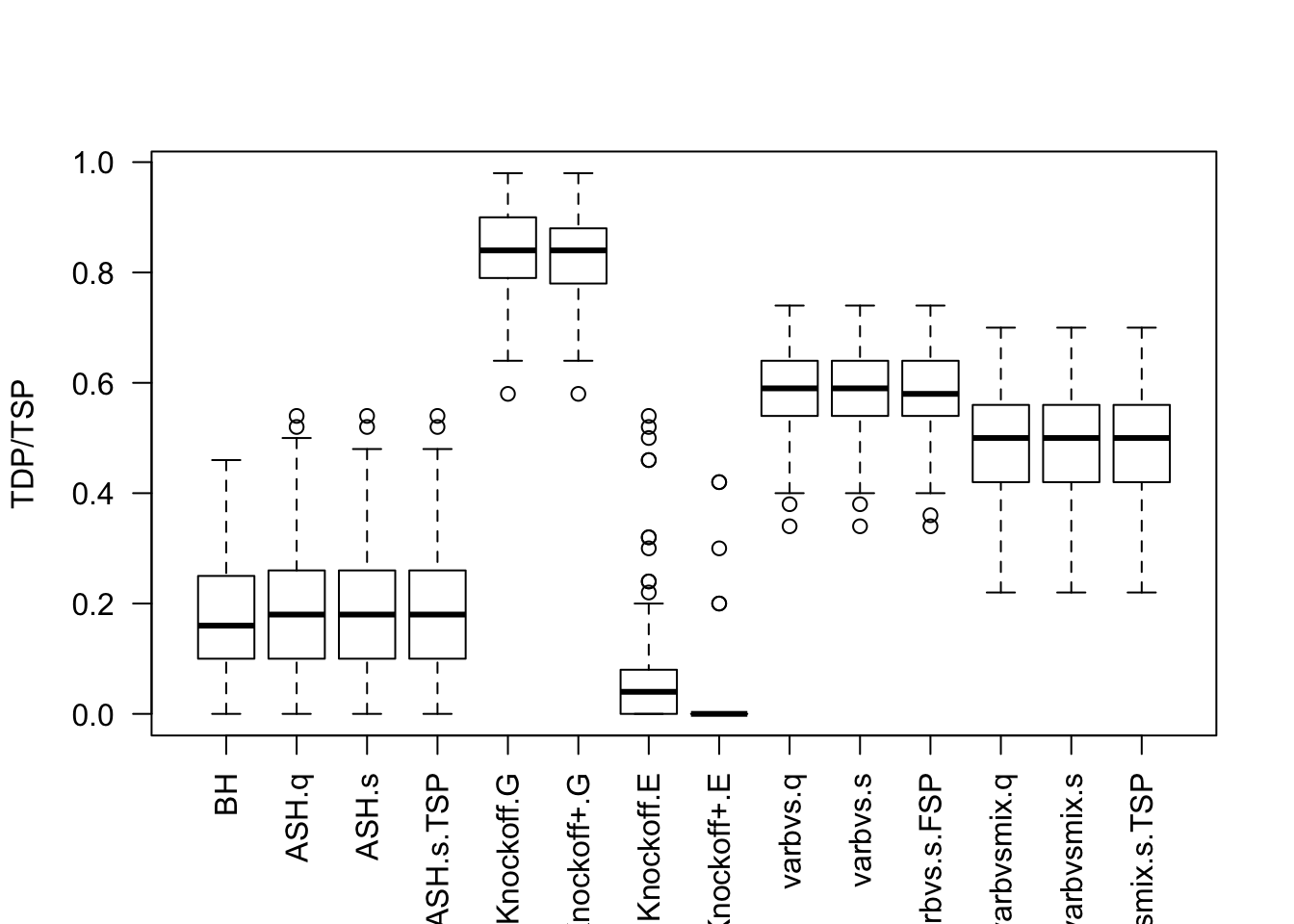
Factor Model for \(\hat\beta\)
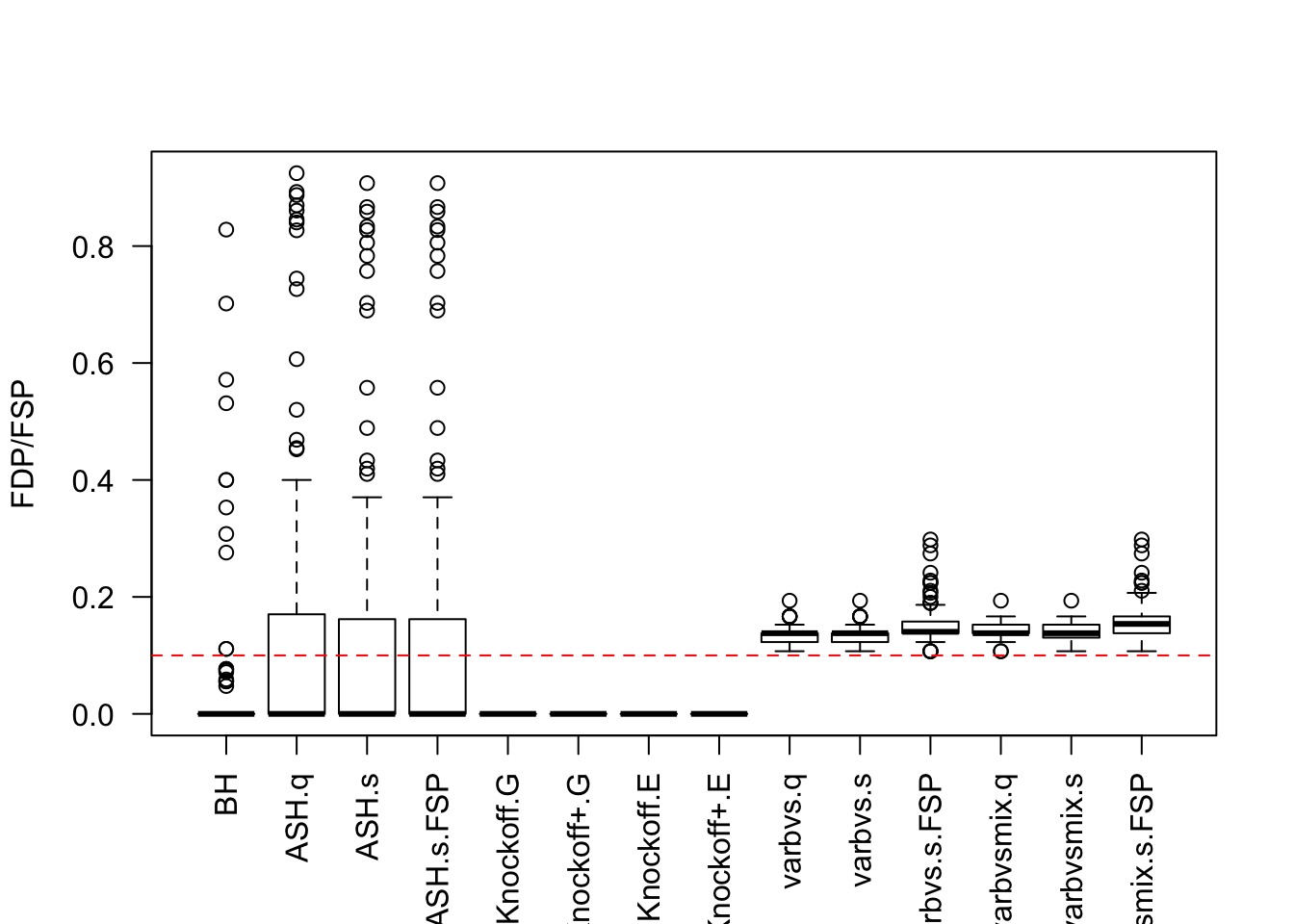
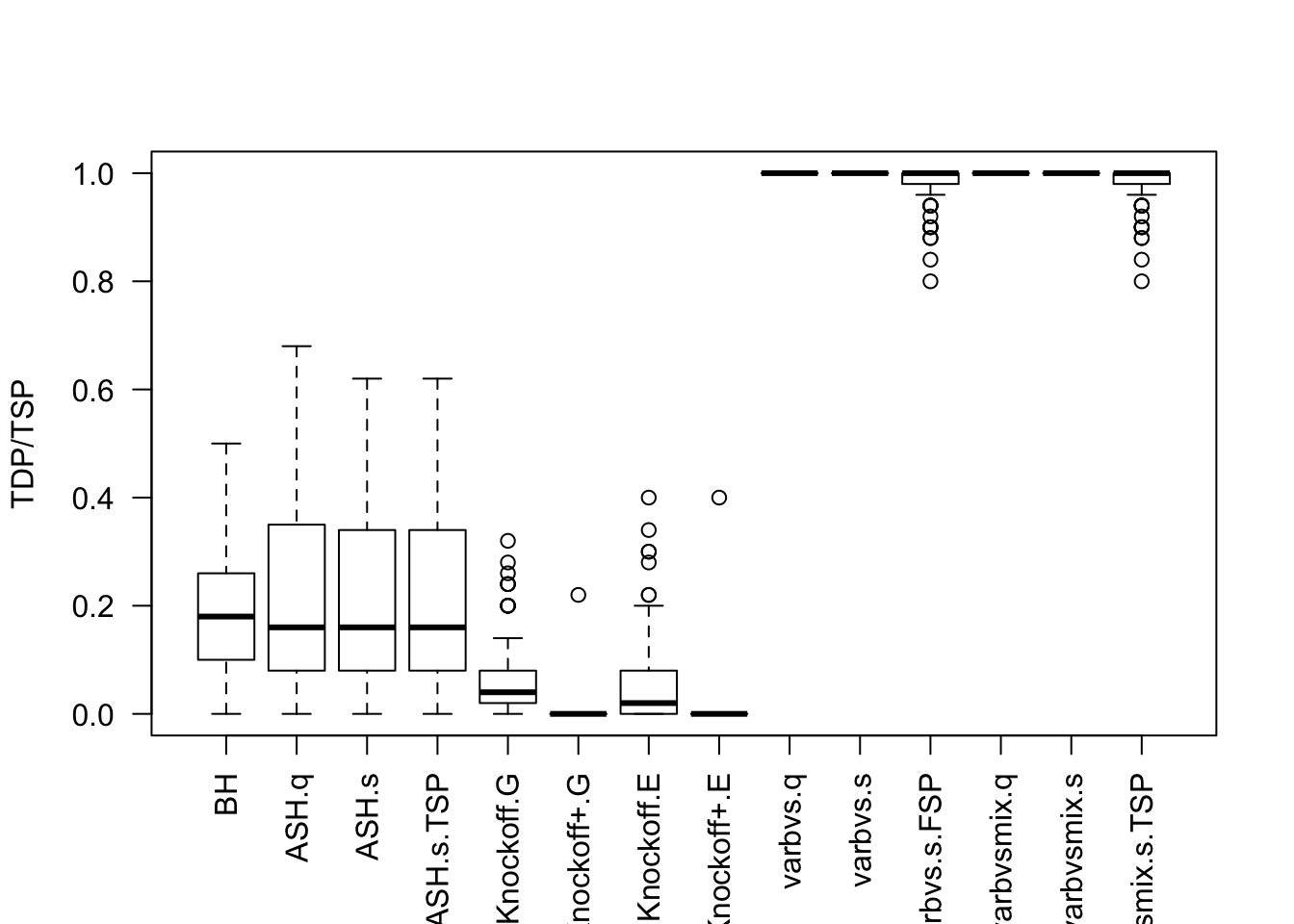
\(n < p\)
n <- 300
p <- 1000
k <- 50
m <- 100
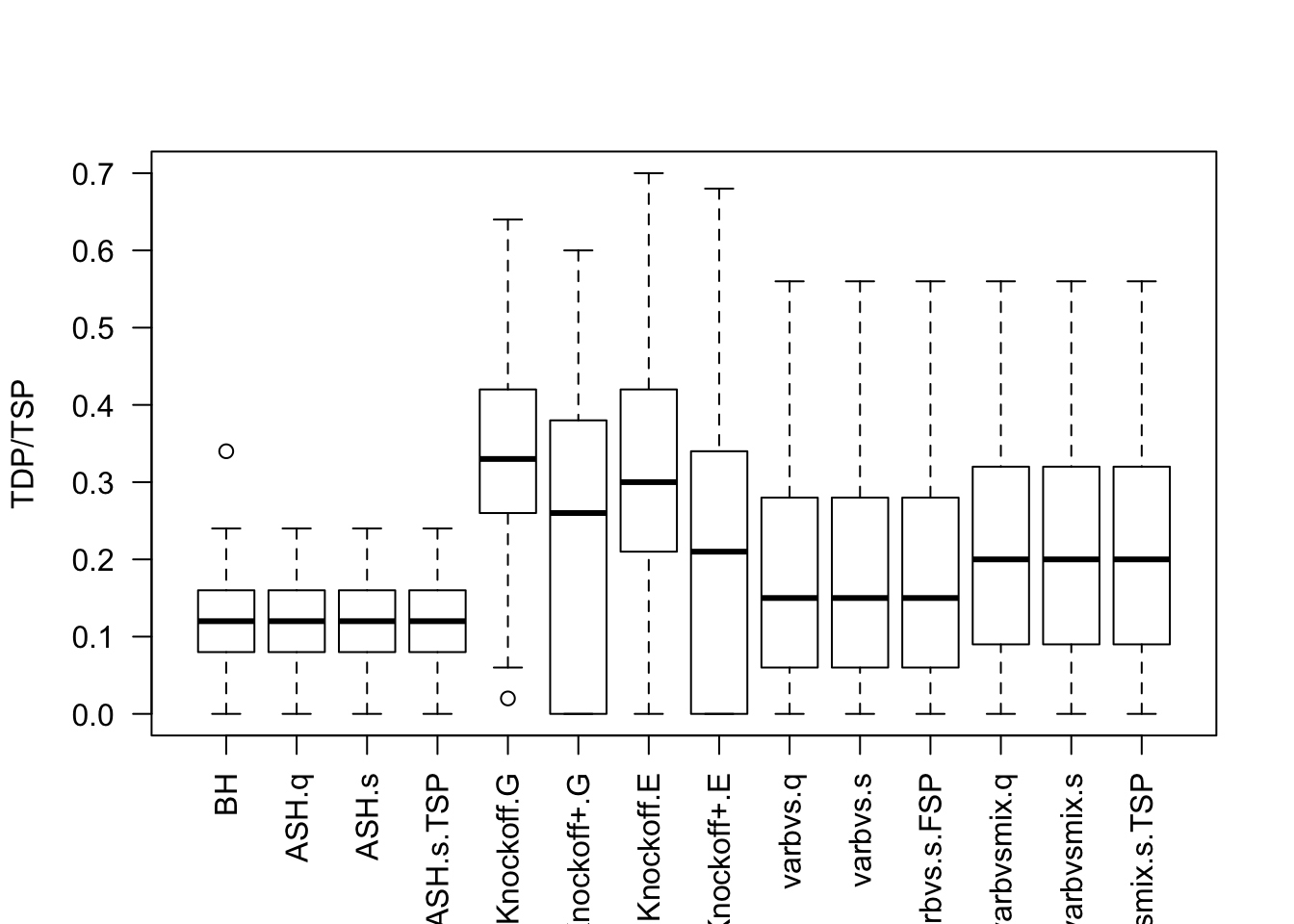
q <- 0.1Independent design
Cov.X <- diag(1 / n, p)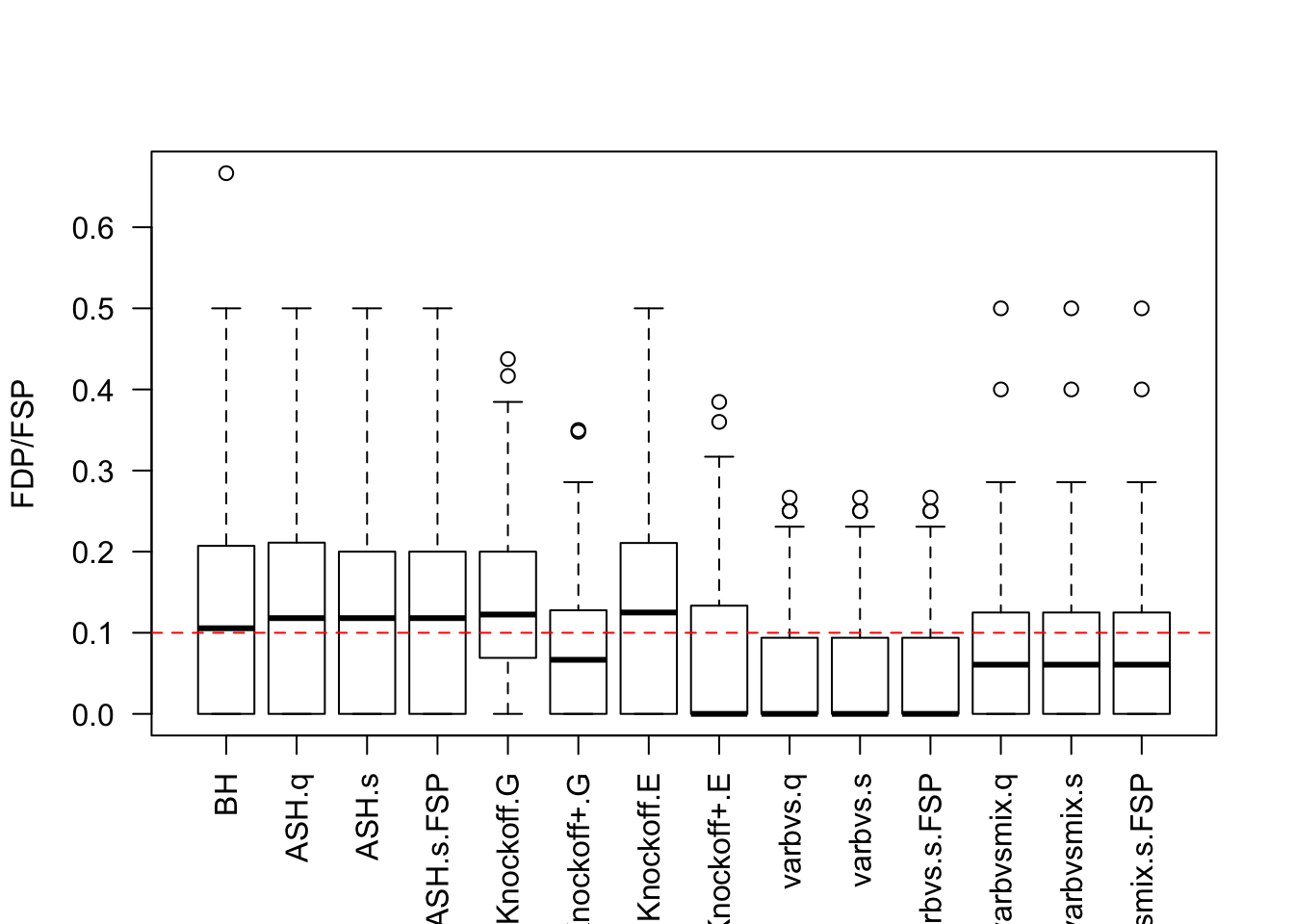
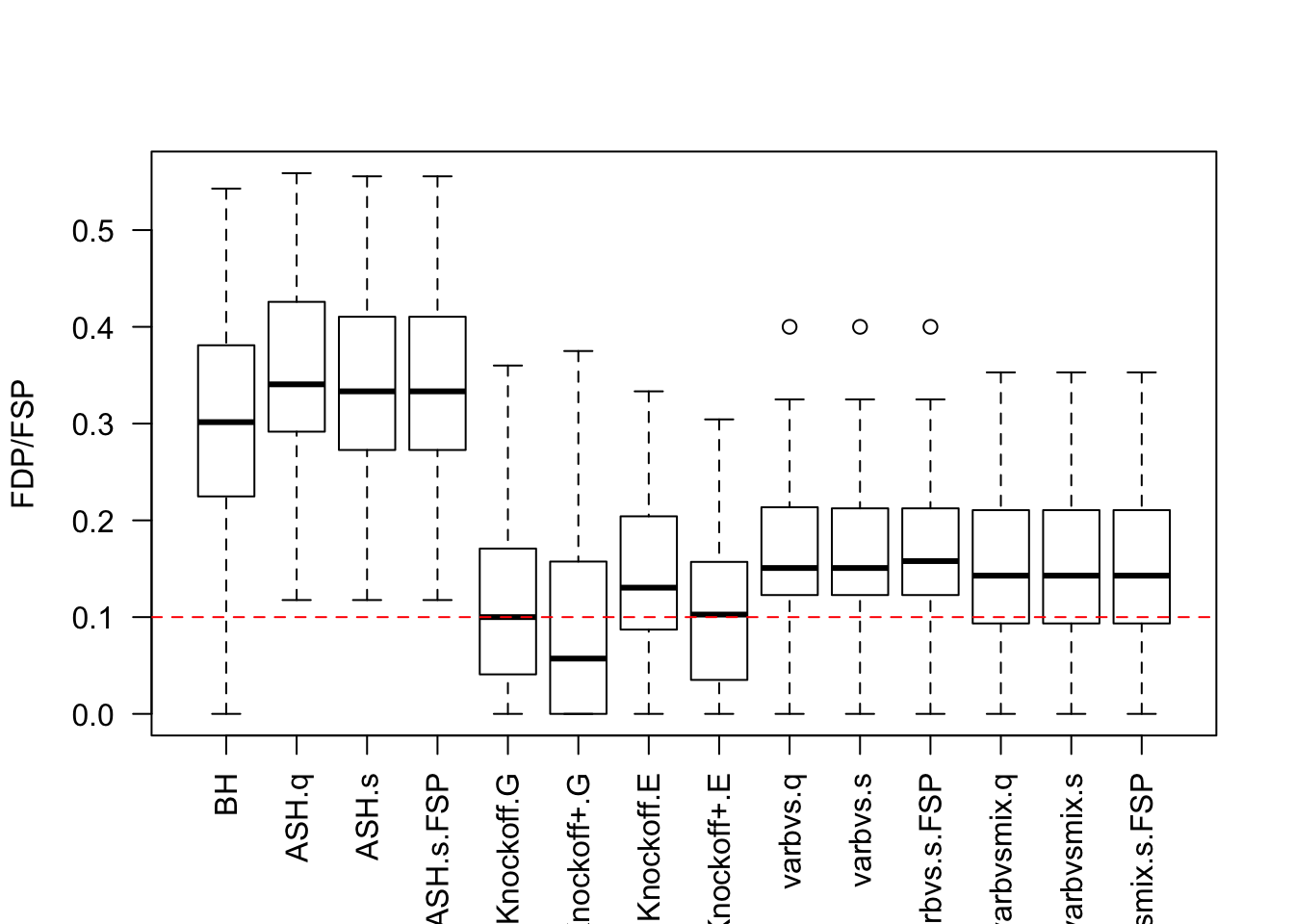
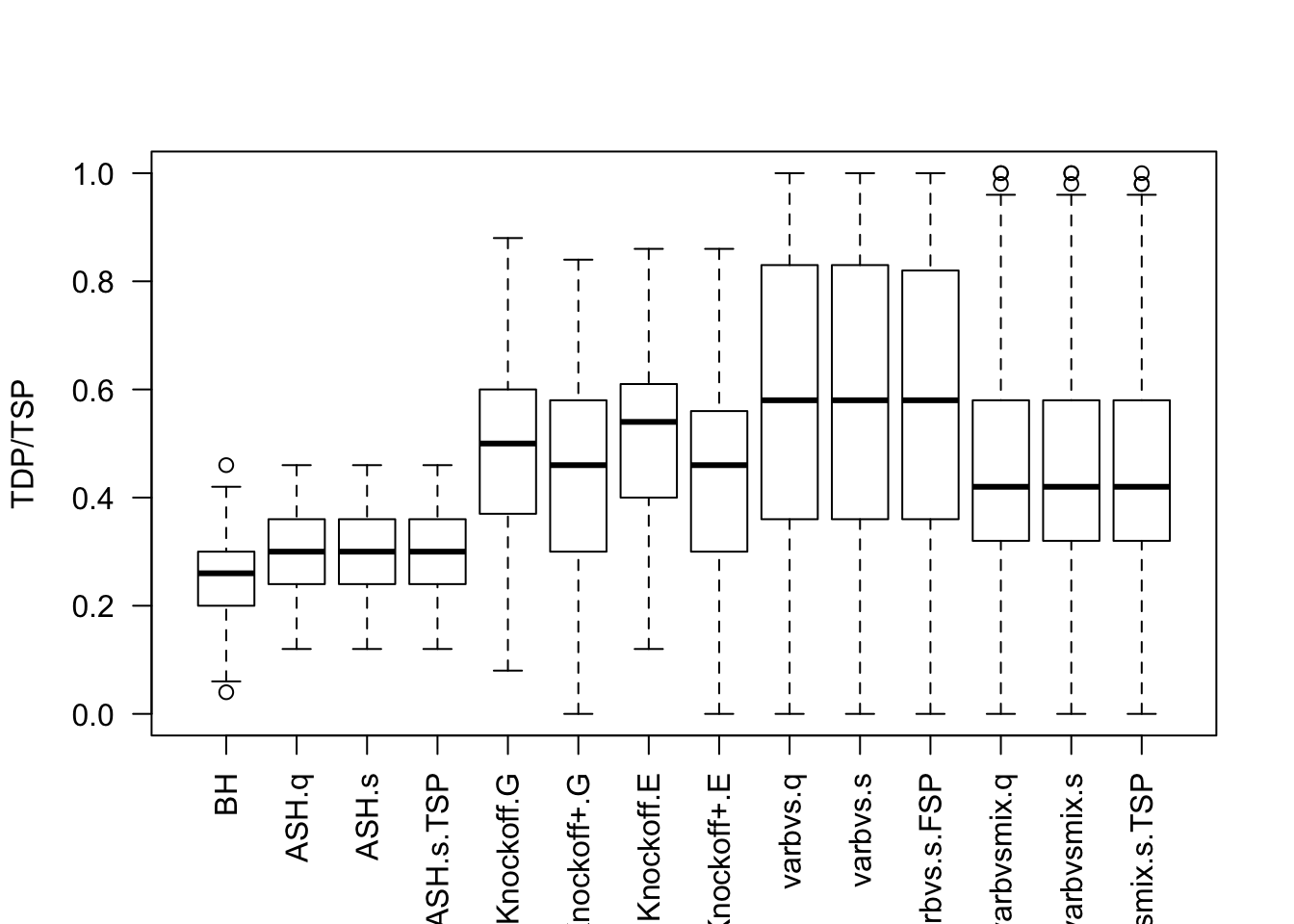
Local correlation / AR model for \(X\)
Factor Model for \(X\)
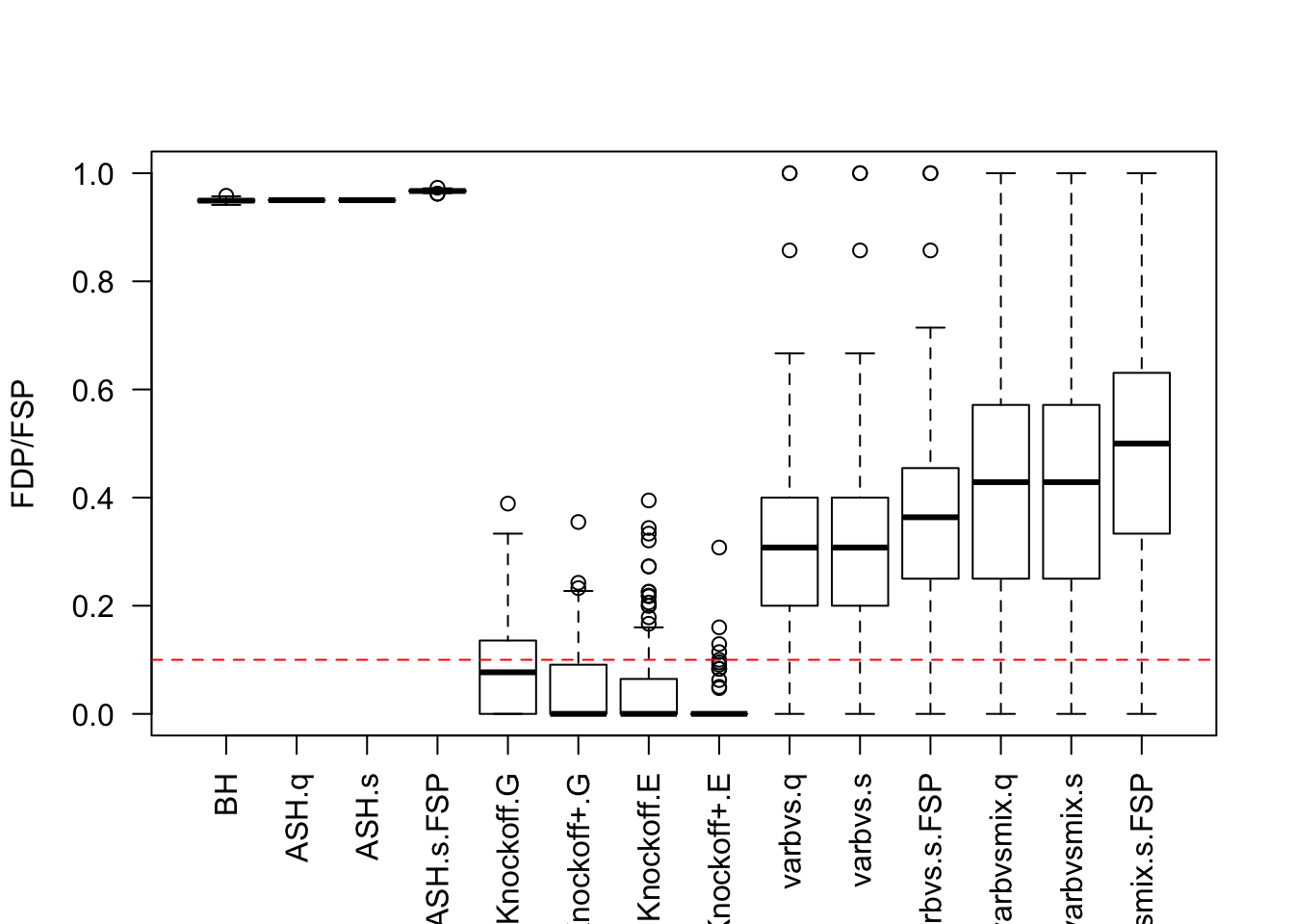
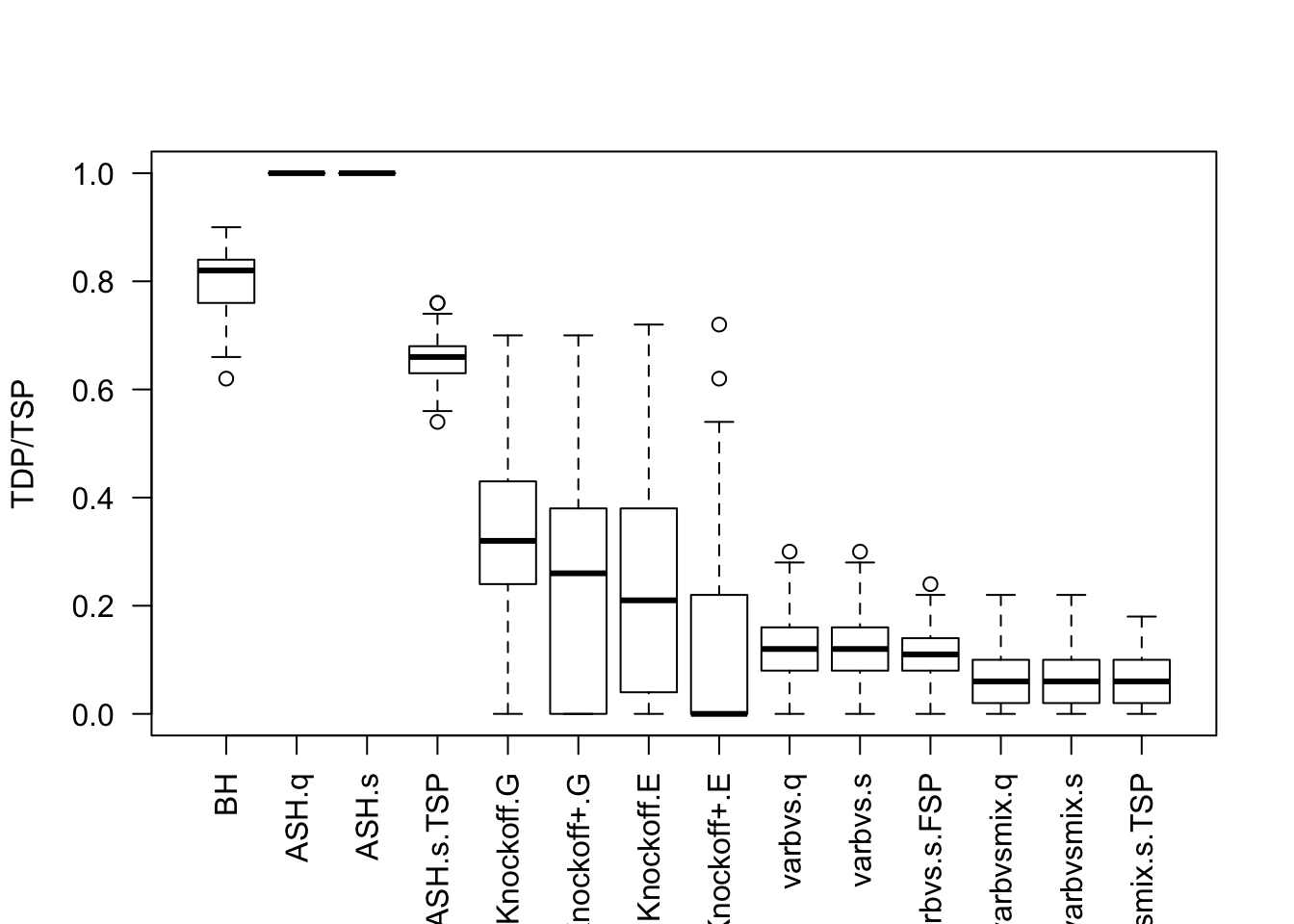
Factor Model for \(\hat\beta\)
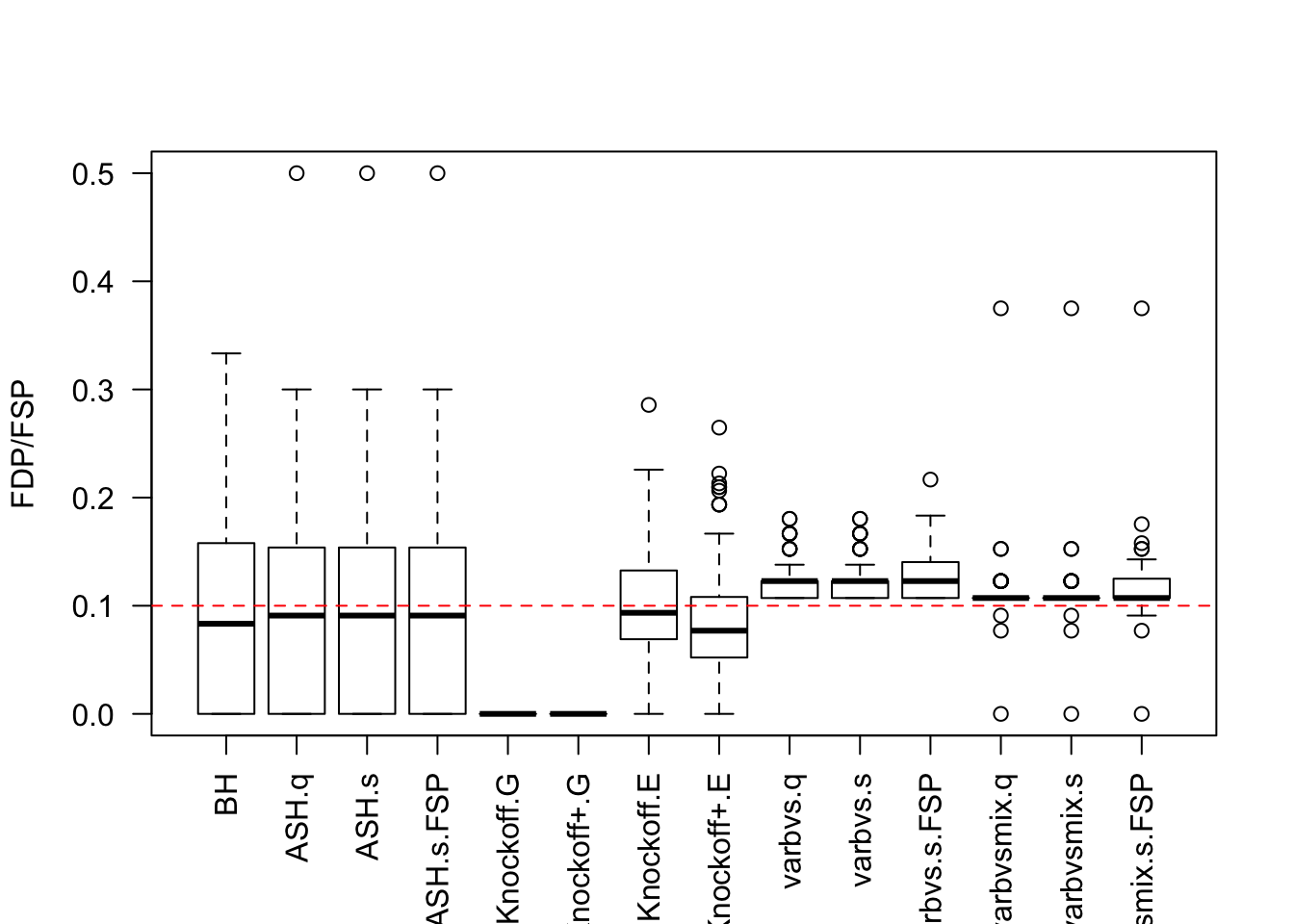
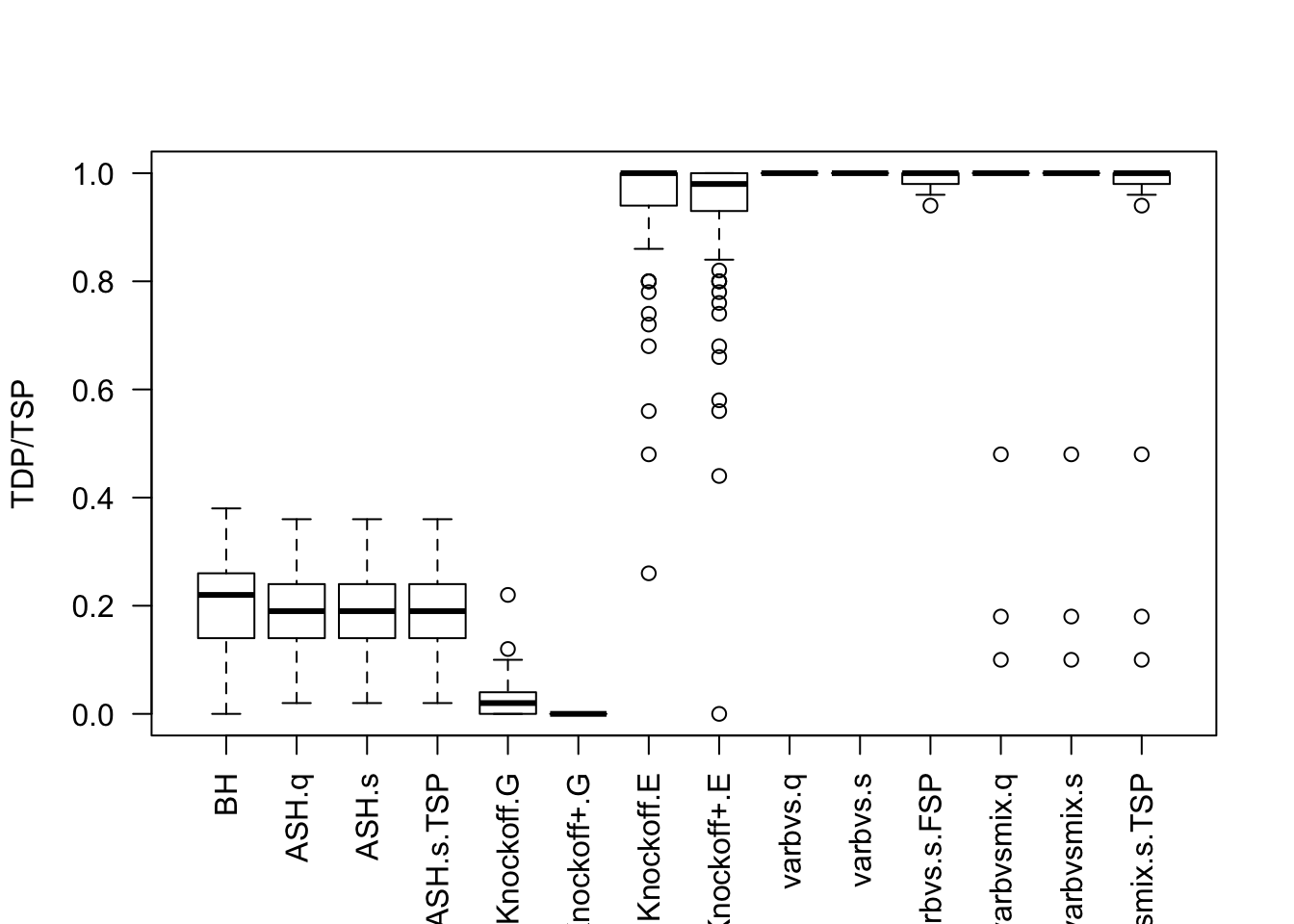
Session information
sessionInfo()R version 3.4.3 (2017-11-30)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS High Sierra 10.13.4
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] parallel stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] lattice_0.20-35 doMC_1.3.5 iterators_1.0.9 foreach_1.4.4
[5] ggplot2_2.2.1 reshape2_1.4.3 Matrix_1.2-12 varbvs_2.5-2
[9] knockoff_0.3.0
loaded via a namespace (and not attached):
[1] Rcpp_0.12.16 knitr_1.20 magrittr_1.5
[4] munsell_0.4.3 colorspace_1.3-2 rlang_0.1.6
[7] nor1mix_1.2-3 stringr_1.3.0 plyr_1.8.4
[10] tools_3.4.3 grid_3.4.3 gtable_0.2.0
[13] latticeExtra_0.6-28 git2r_0.21.0 htmltools_0.3.6
[16] lazyeval_0.2.1 yaml_2.1.18 rprojroot_1.3-2
[19] digest_0.6.15 tibble_1.4.1 RColorBrewer_1.1-2
[22] codetools_0.2-15 evaluate_0.10.1 rmarkdown_1.9
[25] stringi_1.1.6 pillar_1.0.1 compiler_3.4.3
[28] scales_0.5.0 backports_1.1.2 This R Markdown site was created with workflowr