Real Data with Simulated Signals: Part III
Lei Sun
2017-05-20
Last updated: 2017-11-07
Code version: 2c05d59
Introduction
We are comparing our method with ASH and SVA.
library(ashr)
library(edgeR)
library(limma)
library(seqgendiff)
library(sva)
library(pROC)source("../code/gdash.R")Artefactual effects \(\pi_0\delta_0 + \left(1 - \pi_0\right)N\left(0, \sigma^2\right)\) are added to the real GTEx data.
mat = read.csv("../data/liver.csv")We are using \(10K\) genes, \(5\) v \(5\) experimental design, and \(100\) independent simulation trials.
ngene = 1e4
nsamp = 10
N = 100Setting 1: \(\pi_0 = 0.9\), \(\sigma^2 = 1\)
pi0 = 0.9
sd = 1
set.seed(777)
system.time(gdash.comp <- N_simulations(N, mat, nsamp, ngene, pi0, sd)) user system elapsed
18421.805 1175.855 3889.120 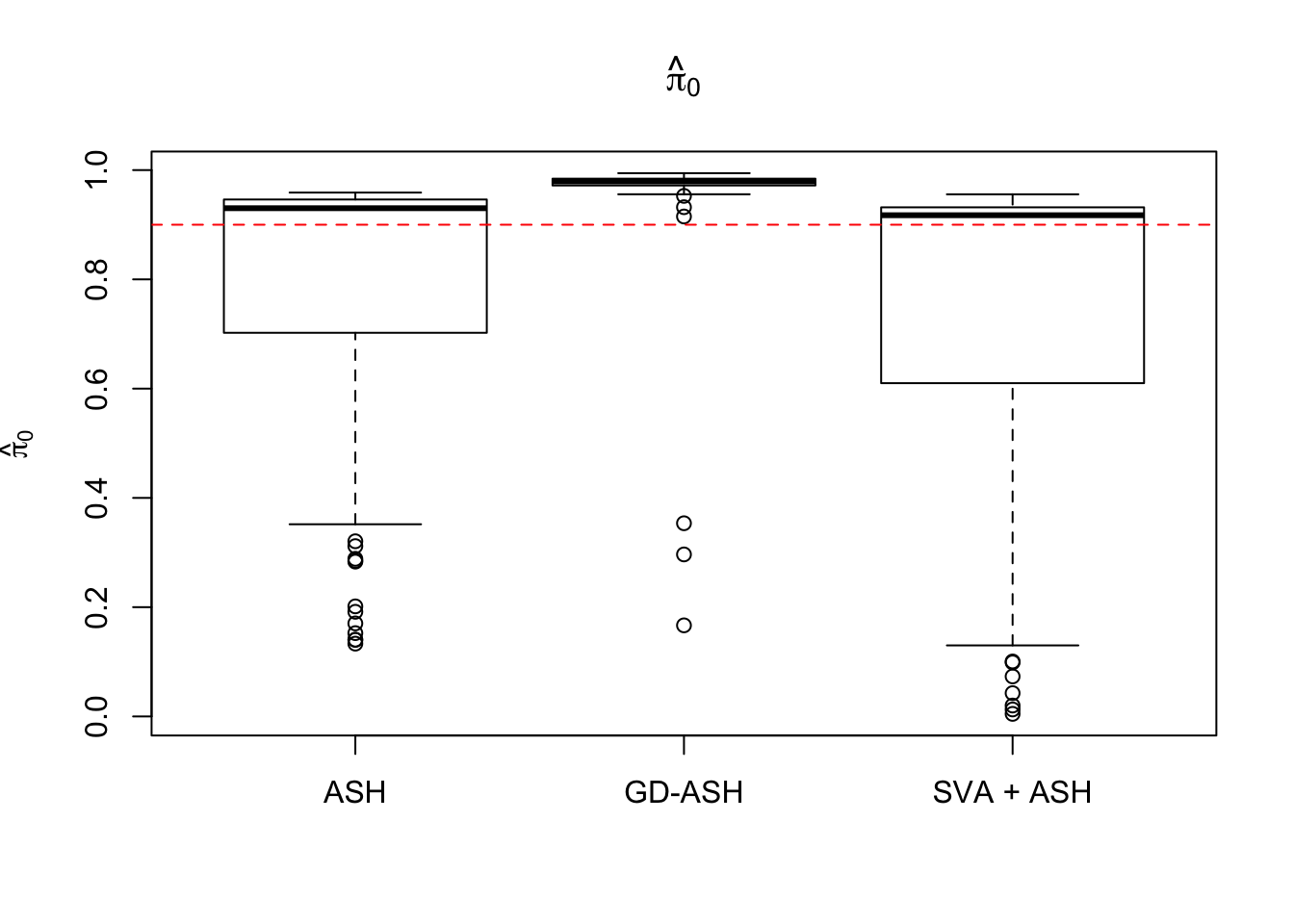
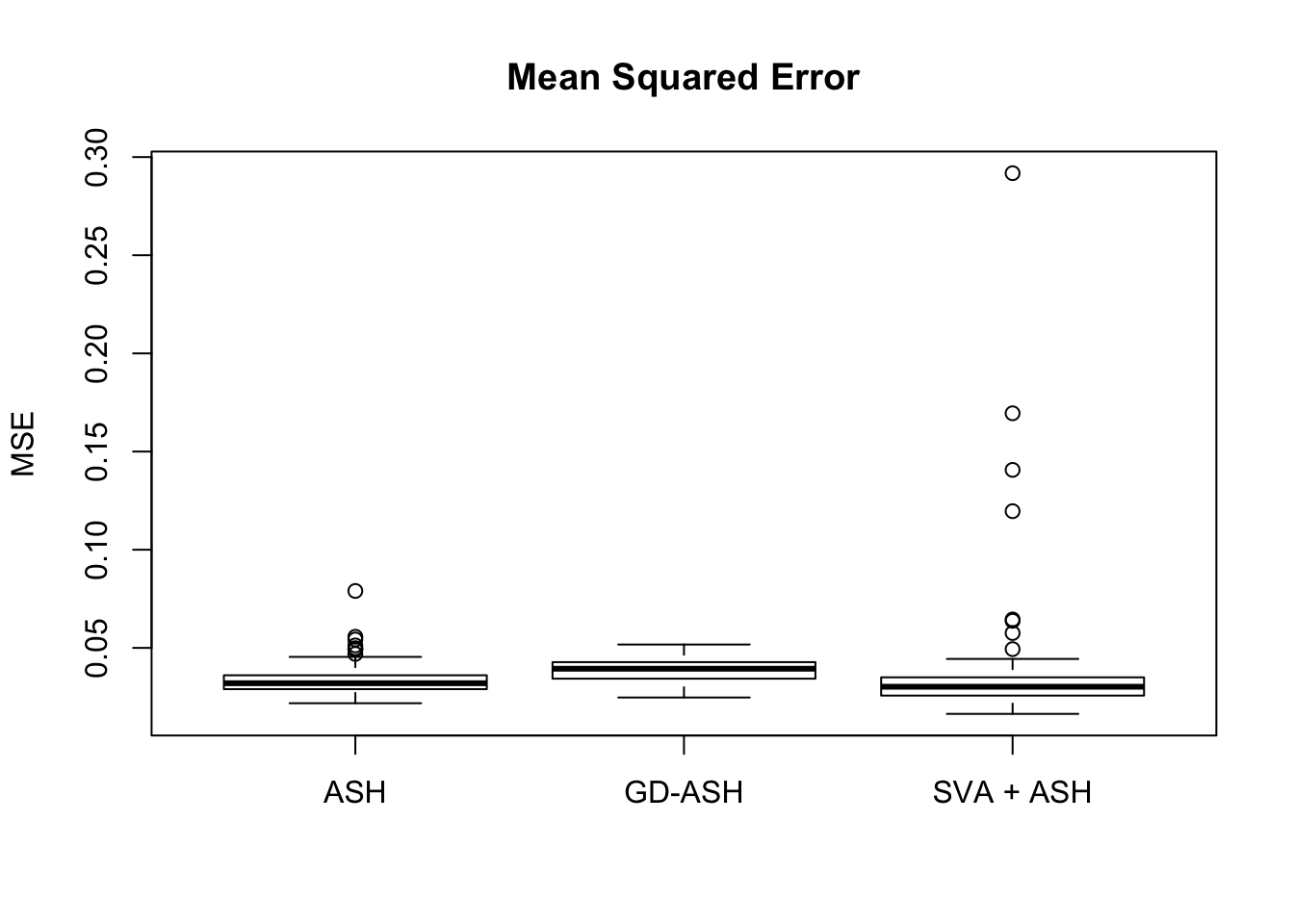
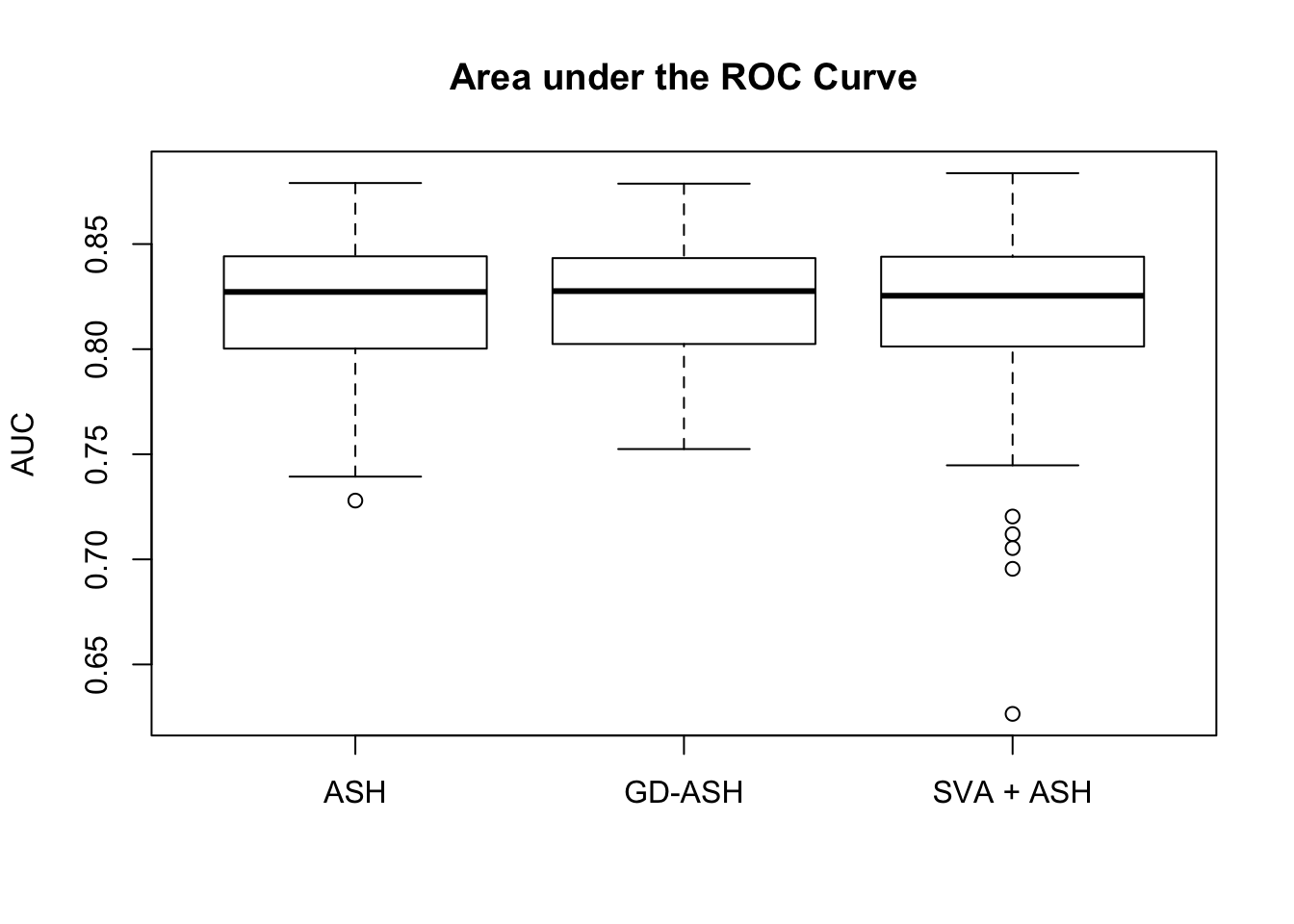
Setting 2: \(\pi_0 = 0.9\), \(\sigma^2 = 2\)
pi0 = 0.9
sd = sqrt(2)
set.seed(777)
system.time(gdash.comp <- N_simulations(N, mat, nsamp, ngene, pi0, sd)) user system elapsed
19523.364 1271.100 4140.141 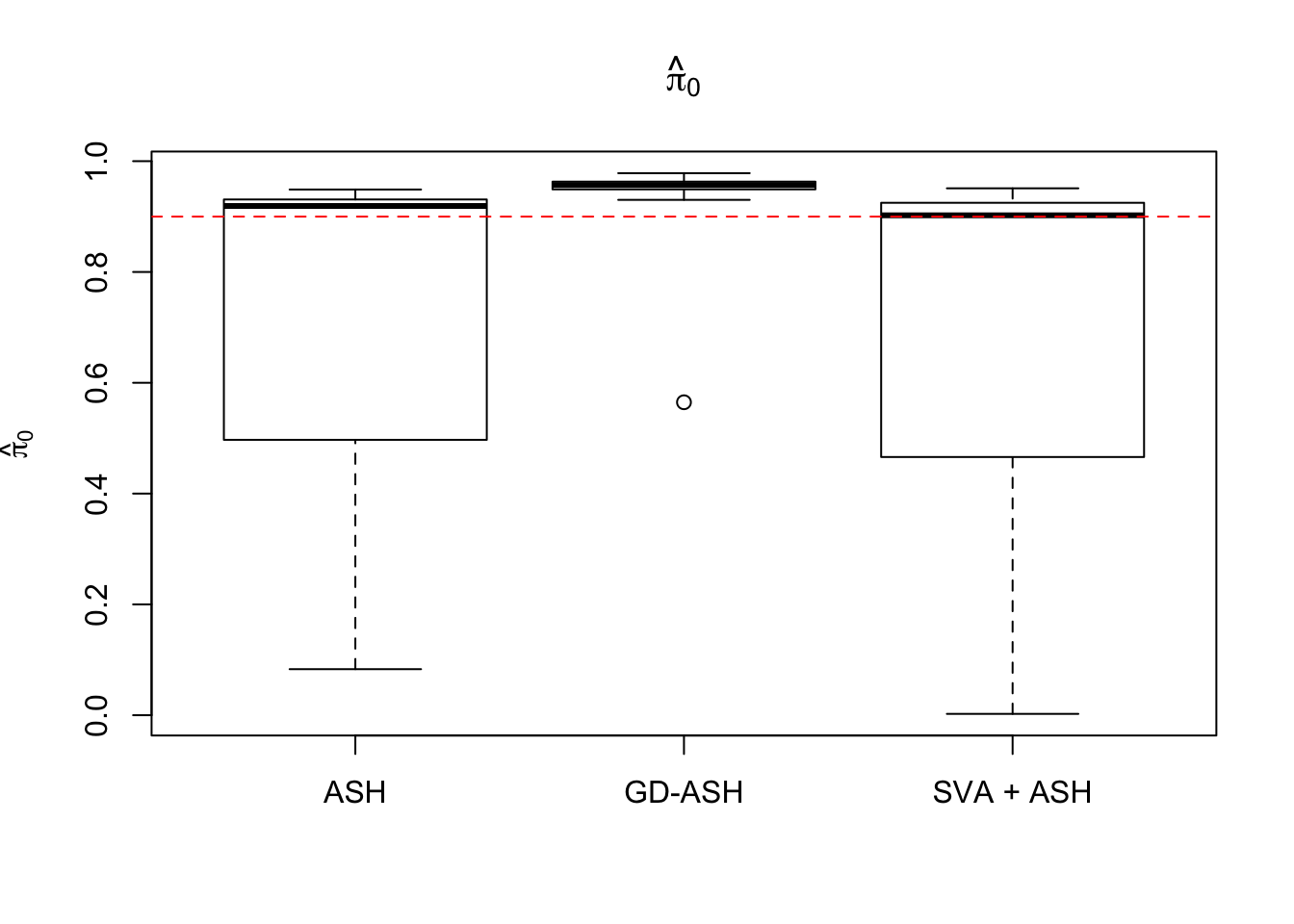
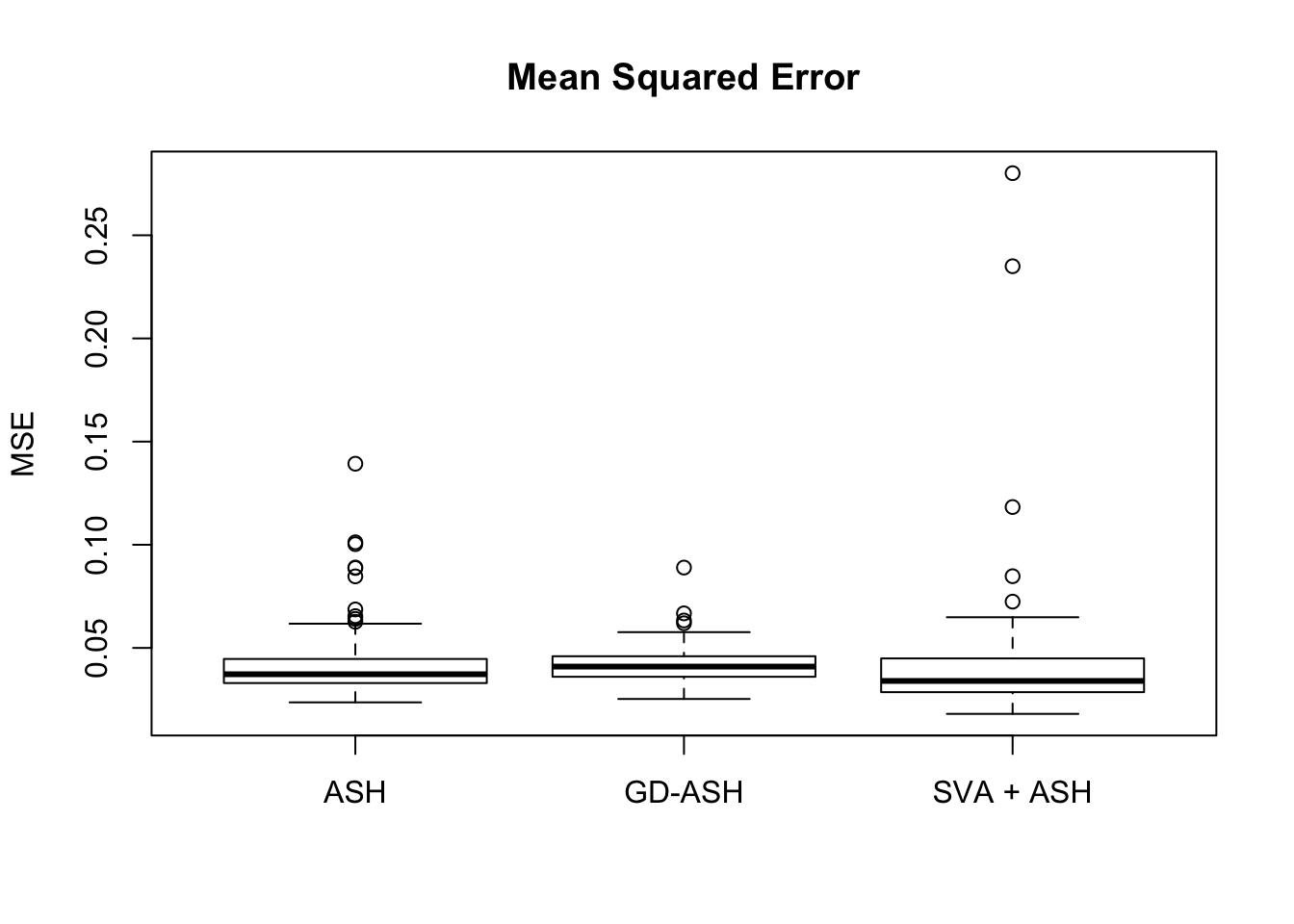
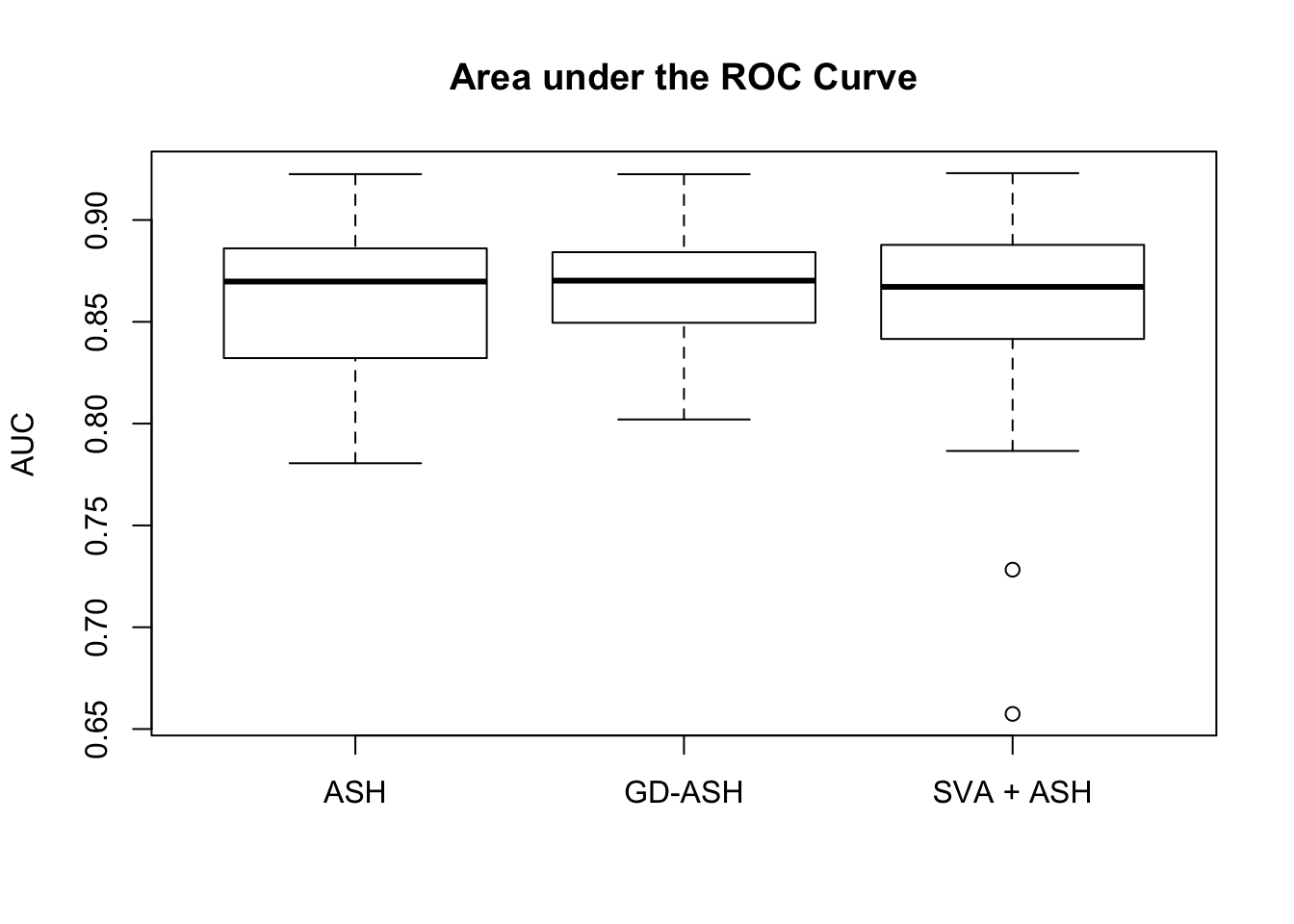
Setting 3: \(\pi_0 = 0.9\), \(\sigma^2 = 3\)
pi0 = 0.9
sd = sqrt(3)
set.seed(777)
system.time(gdash.comp <- N_simulations(N, mat, nsamp, ngene, pi0, sd)) user system elapsed
17770.699 1169.506 3854.622 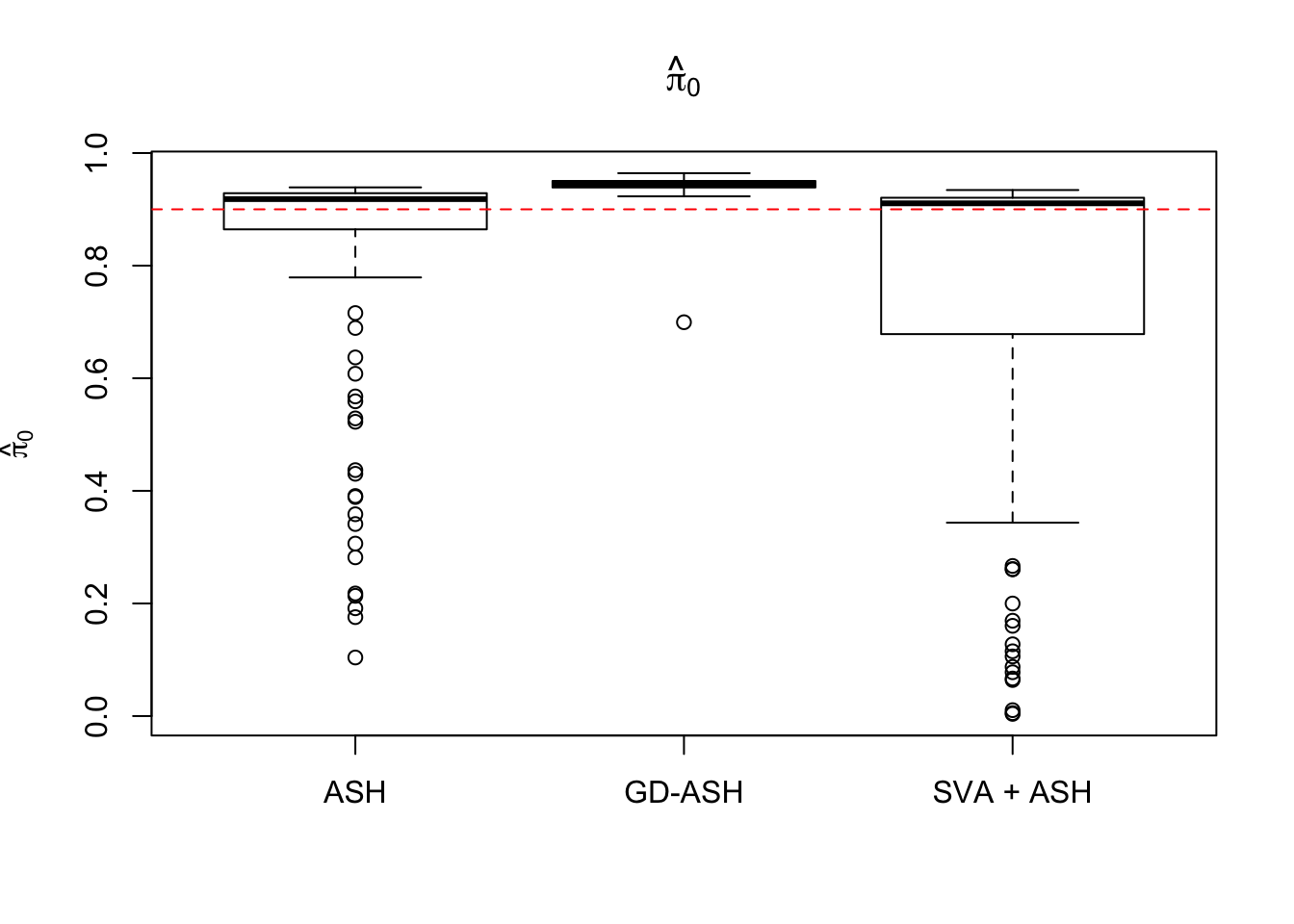
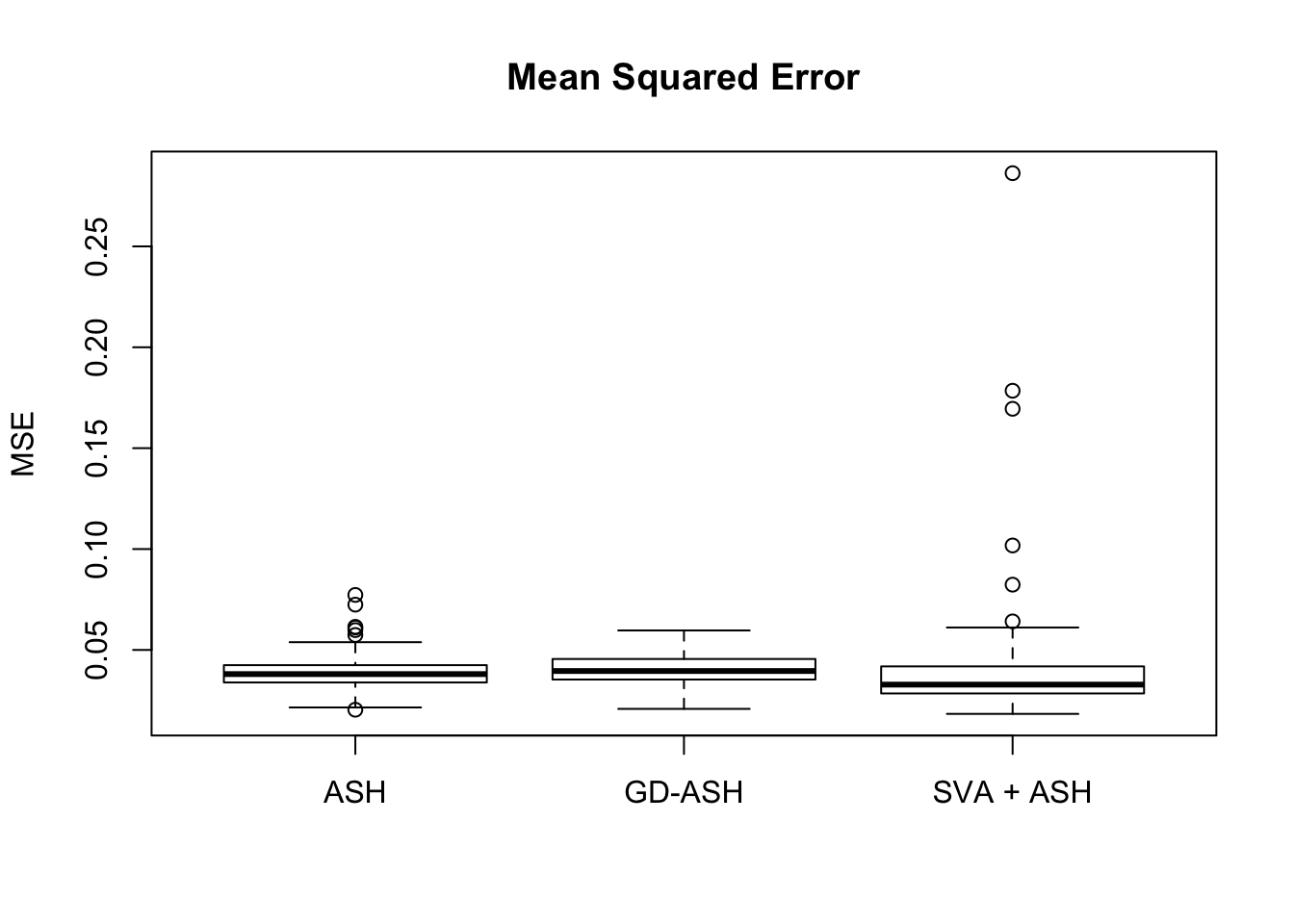
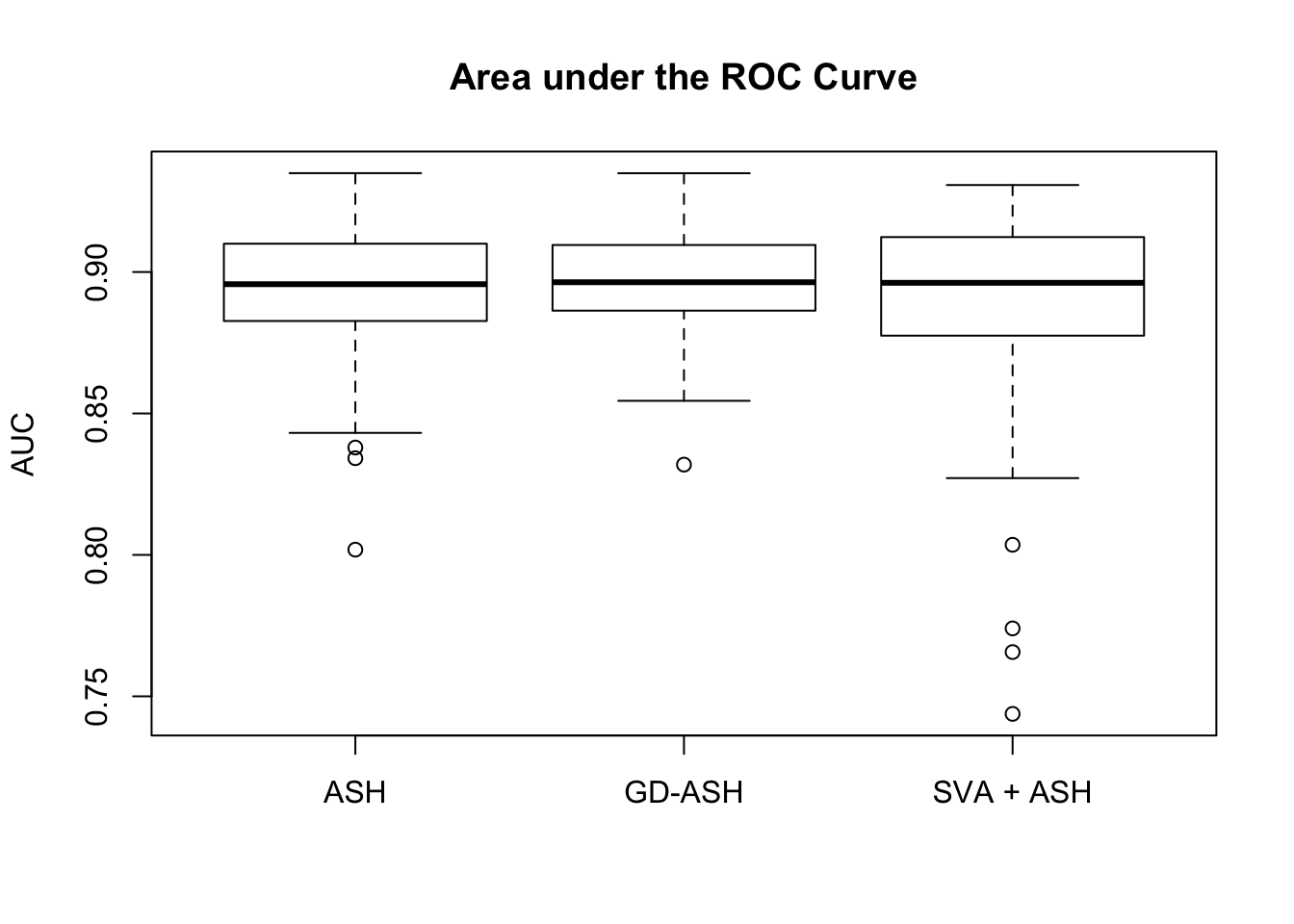
Session information
sessionInfo()R version 3.4.2 (2017-09-28)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS Sierra 10.12.6
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
loaded via a namespace (and not attached):
[1] compiler_3.4.2 backports_1.1.1 magrittr_1.5 rprojroot_1.2
[5] tools_3.4.2 htmltools_0.3.6 yaml_2.1.14 Rcpp_0.12.13
[9] stringi_1.1.5 rmarkdown_1.6 knitr_1.17 git2r_0.19.0
[13] stringr_1.2.0 digest_0.6.12 evaluate_0.10.1This R Markdown site was created with workflowr